International Journal of Diabetes and Clinical Research
African-origin Mitochondrial DNA Variants as a Contributing Factor to Susceptibilities for Diabetes and Age-related Diseases
Kenney MC1,2*, Falatoonzadeh P1, Atilano SR1, Chwa M1, Caceres-del-Carpio J1, Malik D1, Boyer DS3, Nesburn AB1,4 and Kuppermann BD1,5
1Gavin Herbert Eye Institute, University of California Irvine, Irvine, California, USA
2Department of Pathology and Laboratory Medicine, University of California, Irvine, California, USA
3Retina-Vitreous Associates Medical Group, Beverly Hills, California, USA
4Cedars-Sinai Medical Center, Los Angeles, California, USA
5Biomedical Engineering, University of California, Irvine, California, USA
*Corresponding author:
M. Cristina Kenney, MD, PhD, Gavin Herbert Eye Institute, Ophthalmology Research Laboratory, University of California Irvine, 843 Health Science Rd., Hewitt Hall, Room 2028, Irvine, California, 92697, USA, Tel: (949) 824-7603, Fax: (949) 824-9626, Email: mkenney@uci.edu
Int J Diabetes Clin Res, IJDCR-3-053, (Volume 3, Issue 1), Research Article; ISSN: 2377-3634
Received: November 23, 2015 | Accepted: January 11, 2016 | Published: January 13, 2016
Citation: Kenney MC, Falatoonzadeh P, Atilano SR, Chwa M, Caceres-del-Carpio J, et al. (2016) African-origin Mitochondrial DNA Variants as a Contributing Factor to Susceptibilities for Diabetes and Age-related Diseases. Int J Diabetes Clin Res 3:053. 10.23937/2377-3634/1410053
Copyright: © 2016 Kenney MC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
African-origin populations are more susceptible to diabetes and other age-related diseases compared to European-origin populations, but mechanisms for the differential susceptibility remain unknown. Human mitochondrial (mt) DNA haplogroups are maternally inherited ancient polymorphisms representing different geographic origins of populations. Haplogroups are defined by accumulations of single nucleotide polymorphisms (SNPs), some of which cause changes in amino acids and rates of mtDNA replication and transcription. Several studies have shown that age-related diseases and their complications can be linked to mtDNA haplogroup subsets. Previous studies report that transmitochondrial cybrids (cytoplasmic hybrids), which contain identical nuclei but either European-origin (H) or African-origin (L) haplogroup mtDNA, have significantly different bioenergetic profiles, production levels of reactive oxygen species, and expression levels for complement, inflammation and apoptosis genes, thus suggesting that major biological pathways can be modulated by mtDNA. Using GeneChip arrays and Q-PCR, we show that African-origin L cybrids show significantly different expression levels for three Wnt pathway genes (DKK3, SFRP1, and KREMEN1) and three diabetes-related genes (RPS6KA4, ADAMTS9, and VEGFA) compared to European-origin H cybrids. Our current findings, along with others, support the hypothesis that an individual’s mtDNA can modulate the expression of important Wnt signaling and diabetes-related genes, which may contribute to the racial/ethnic disparities associated with diabetes and other age-related diseases.
Keywords
mtDNA, Wnt pathway/β-catenin, Disease Susceptibilities, Diabetes, African-origin population, European-origin population
Abbreviations
ABI: Applied Biosystems; ADAMTS9: ADAM metallopeptidase with thrombospondin type 1 motif, 9; ARPE-19: Retinal pigmented epithelium cell line; ATP: Adenosine triphosphate; CSNK1E: Casein kinase 1, epsilon; DKK3: Dickkopf 3 homolog; DMEM: Dulbecco’s modified Eagle’s medium; DNA: Deoxyribonucleic Acid; GSK3A: Glycogen synthase kinase 3 alpha; KREMEN1: Kringle containing transmembrane protein 1; μl: Microliter; ng: Nanogram; OXPHOS: Oxidative phosphorylation; Q-PCR: Quantitative polymerase chain reaction; PCR: Polymerase chain reaction; RARA1: Retinoic acid receptor, alpha; RPS6KA4: Ribosomal protein S6 kinase, 90kDa, polypeptide 4; SEM: Standard error of the mean; SNPs: Single nucleotide polymorphisms; SFRP1: Secreted frizzled-related protein 1; UCLA: University of California, Los Angeles; VEGFA: Vascular endothelial growth factor A.
Introduction
All mitochondria (mt) contain maternally inherited, circular, double-stranded DNA consisting of 16,569 nucleotide pairs. The coding region of mtDNA encodes for 37 genes including 13 protein subunits essential for OXPHOS, 2 ribosomal RNAs and 22 transfer RNAs [1-3]. The non-coding mtDNA D-loop region is important for replication and transcription. Recently evidence shows that mtDNA can also code for small, biologically active “Mitochondrial Derived Peptides” [4,5] supporting a more robust role for the mitochondrial genome in diseases.
Human mtDNA haplogroups are ancient polymorphisms inherited along maternal lineages and have been used for large ancestral population studies. These adaptive single nucleotide polymorphism (SNP) changes have occurred over thousands of years and define mtDNA backgrounds connected to geographic origins of human populations. There are eight different European haplogroups with the most common being the H haplogroup (approximately 30%, www.MitoMap.org). Individuals of maternal African-origin belong to the L haplogroups and being the oldest haplogroups, they contain the largest diversity of SNPs.
Some racial/ethnic populations are more prone to develop specific diseases [6-11]. The incidence of diabetes is highest in the non-Hispanic black population with the non-Hispanic white adults having the lowest risk (http://www.diabetes.niddk.nih.gov/dm/pubs/statistics). Alzheimer’s disease (AD) is another example where the prevalence is associated with the racial/ethnic background of the individual. Compared to the non-Hispanic white population, the African-Americans are twice as likely to develop AD or dementia [12-14] and also have higher incidence of systemic lupus erythematosus (SLE) [10,11]. The causes for these racial/ethnic differences are not known but epigenetic changes may play a role [15]. However, there is mounting evidence that mtDNA haplogroups, which define different racial populations, may contribute to differences in disease susceptibilities [16].
The majority of studies related to genetics and diseases have focused on the contribution of nuclear genotypes with many fewer studies on the mtDNA genome. One reason has been the difficulty to assign functional, cellular changes to the mtDNA variants. However, with the development of the transmitochondrial cybrid (cytoplasmic hybrid) model, many questions related to the functional importance of mtDNA haplogroup variants and mitochondrial-nuclear interactions can be addressed. Cybrid studies comparing European-origin haplogroups (H and J) and African-origin haplogroups (L) showed differences in cell growth, oxygen consumption rates, expression of non-energy-related genes, responses to stressors, and rates of glycolysis [16-20]. Based upon many differences in the cellular behavior in the H and L cybrids, we hypothesized that the cybrids might also have different gene expression patterns for Wnt/β-catenin signaling genes, which are important for cell proliferation, cell migration, apoptosis, development and numerous diseases, including lipid metabolism, glucose homeostasis, diabetes, Alzheimer’s disease and cancers [21-23].
Our findings demonstrate that cybrids containing African-origin mtDNA (L haplogroup) have significantly different expression levels of three Wnt pathway genes (DKK3, SFRP1, and KREMEN1) and three diabetes-related genes (RPS6KA4, ADAMTS9, and VEGFA) compared to the European-origin H haplogroup cybrids. analyses of the Functional Pathways on the GeneChip array showed differences between L and H cybrids for development and cancer systems. Our current findings suggest that subjects with African-origin mtDNA versus European - origin mtDNA may have different expression levels of Wnt signaling and diabetes-related genes, which thereby may contribute to the racial/ethnic disparities associated with diabetes, cancers and Alzheimer’s diseases.
Materials and Methods
Transmitochondrial cybrids and culture conditions
Institutional review board approval was obtained from the University of California, Irvine (#2003-3131). For DNA analyses, 10mls of peripheral blood were collected via venipuncture in tubes containing 10mM EDTA from normal young volunteers. DNA was isolated with a DNA extraction kit (PUREGENE, Qiagen, Valencia and CA). Platelets were isolated by a series of centrifugation steps and final pellets were suspended in Tris-buffered saline. The ARPE-19 cells deficient in mtDNA (Rho0) were created by serial passage in low dose ethidium bromide [24]. Cybrids were produced by polyethylene glycol fusion of platelets with Rho0 ARPE-19 cells according to modified procedures of Chomyn [25]. All experiments used passage 5 cybrid cells for the assays described below.
Identification of cybrid haplogroups
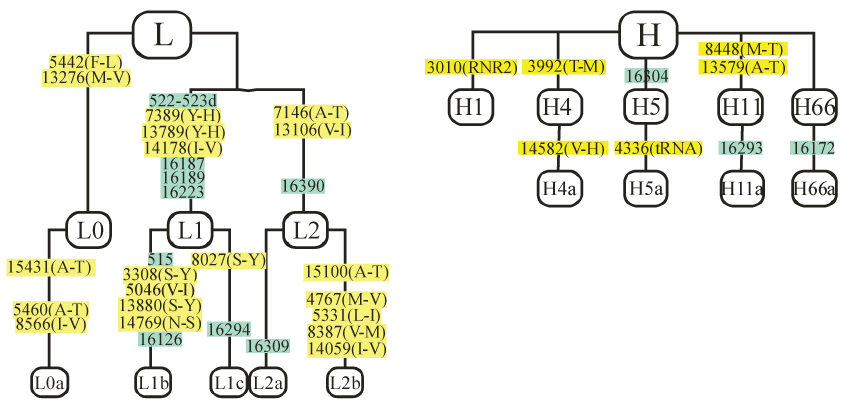
Verification of transfer of the mitochondria into the Rho0 ARPE-19 cells was accomplished by polymerase chain reaction (PCR), restriction enzyme digestion, or sequencing of the mtDNA to identify the mitochondrial haplogroup of each cybrid. Briefly, cybrid DNA was extracted from cell pellets using a spin column kit (DNeasy Blood and Tissue Kit, Qiagen) and quantified using the Nanodrop 1000 (Thermo Scientific, Wilmington, DE). PCR and restriction enzyme digests [26], allelic discrimination and sequencing were performed to determine mitochondrial haplogroups. The primers for allelic discrimination were synthesized by ABI Assay-by-Design. The samples were run at GenoSeq, the UCLA Genotyping and Sequencing Core, on an ABI 7900HT. Data were analyzed with Sequence Detection Systems software from ABI. The SNPs defining the H haplogroup were T7028C and G73A. Further subtyping showed the H cybrids were H5a (n = 1), H66a (n = 1), H11a (n =1), H4a (n = 1) and H (n = 4). The L cybrids were further subtyped to be L0a (n = 1), L1b (n = 1), L1c (n = 1), L2a (n = 2) and L2b (n = 1) defined by the SNP variants (Figure 1).
.
Figure 1: Branching diagram of L haplogroups subsets showing the different SNPs that define the specific haplogroup. The yellow highlighted SNPs are non-synonymous and cause amino acid changes in the coding regions of the mtDNA (5 amino acid changes for L0a; 7 amino acid changes for the L1b and L2b individuals, 2 amino acid changes for L2a and 4 amino acid changes for L1c, respectively) compared to the European H haplogroup (Cambridge reference). The green highlighted SNPs occur in the Control Region (non-coding MT-Dloop), which is critical for replication and transcription. The L haplogroups, which represent older geographic origins, contain higher numbers of non-synonymous SNPs than the European haplogroups and these differences may contribute to the racial/ethnic susceptibilities associated with diseases. MT, mitochondrial; SNP, single nucleotide polymorphism.
View Figure 1
Analyses of GeneChip arrays and statistical analyses
For the GeneChip array analyses, equal amounts of RNAs (250 ng/μl per sample) from either H cybrids (n = 3) or L cybrids (n = 3) were combined into two separate samples. The two separate RNA samples were sent to the UCLA Clinical MicroArray Core Lab for analyses with the Affymetrix Human U133 Plus 2.0 Array data uploaded at Gene Expression Omnibus. Gene expression results were analyzed with IPA summary pathway analyses software (INGENUITY Systems, Redwood City, CA).
Quantitative polymerase chain reaction (Q-PCR) analyses
Gene expression changes identified by the GeneCHip array were verified by Q-PCR using 12 different primers (QuantiTect Primer Assay, Qiagen) for genes associated with the Wnt signaling or diabetes-related pathways. The Q-PCR analyses were performed on individual H or L cybrids and not on combined samples. Total RNA was isolated from individual pellets of cultured cells of haplogroup H cybrids (n = 8) and haplogroup L cybrids (n = 6) as described above. Q-PCR was performed using a QuantiFast SYBR Green PCR Kit (Qiagen) on a Bio-Rad iCycleiQ5 detection system. Analyses were performed in triplicate. Statistical analyses of gene expression levels were performed to measure differences between cybrids groups using Prism, version 5.0; (Graph Pad Software Inc.). Fold values were calculated using the standard 2^ddCt formula (ddCt = delta delta Ct). The H Cybrids were used as the relative control for L cybrids. The P-value < 0.05 was considered statistically significant.
Results
GeneChip array analyses
The pooled RNA isolated from H cybrids or L cybrids were analyzed using the GeneChip array that characterizes expression levels for over 40K genes. The IPA summary program of the GeneChip array showed that the pooled RNA from the L cybrids had differences in the Wnt/β-catenin signaling pathway compared to the pooled RNA from the H cybrids. More detailed analyses by the Ingenuity program shows that in the L cybrids, SFRP1 gene had at least 2-fold lower expression levels while the KREMEN1, RARA, and ADAMST9 genes showed greater than 2-fold increase when compared to the H cybrids (Table 1).
![]()
Table 1: Genes expressed in WNT/β-catenin signaling pathways for L cybrids versus H cybrids as determined by IPA summary program analyses of Affymetrix Human U133 Plus 2.0 Array.
View Table 1
With the GeneChip array analyses, we were surprised by the differences in the Wnt/β-catenin signaling genes expression because all cybrids had the identical nuclei, cytoplasm and culture conditions but varied only in the mtDNA haplogroup of their mitochondria. This finding is important because there are many Functional Pathways that use the Wnt signaling pathway (Table 2). When the Functional Pathways were compared for the European-origin H cybrids versus the African-origin L cybrids, there were thirteen categories different between the two groups, including cancer, developmental, reproductive, cellular movement and morphology. These differences can be assigned to influences of the mtDNA variants since the nuclear genomes within all cybrids are identical.
![]()
Table 2: Description of Functional Pathways Regulated by Wnt/β-catenin Signaling Pathway as Determined by IPA Summary Program analyses of Affymetrix Human U133 Plus 2.0 Array.
View Table 2
Q-PCR analyses
To verify the GeneChip array results, we performed Q-PCR on individual RNA samples from the H (n = 8) or L (n = 6) cybrids for twelve nuclear genes associated with the Wnt/β-catenin signaling or diabetes-related pathways. We examined genes that showed greater than 2-fold differences in the H and L cybrids or genes that were critical for the Wnt pathway. Table 3 depicts GenBank Accession numbers and a brief description of the functions for nine genes associated with the Wnt/β-catenin signaling pathway (CSNK1E, DKK3, SFRP1, KREMEN1, WNT9, GSK3A, RARA1, LRP1 and MDM2), and three genes associated with diabetes (RPS6KA4, ADAMTS9 and VEGFA) [27].
![]()
Table 3: a. Description of Wnt/β-catenin Genes Analyzed by Q-PCR; b. Description of diabetes-related genes analyzed by Q-PCR.
View Table 3
Although all cybrids possess identical nuclei and culture conditions, the L cybrids had different expression levels for three Wnt-related genes compared to H cybrids (Table 4). Expression levels for DKK3 (0.06-fold, P = 0.03) and SFRP1 (0.5-fold, P = 0.003) were decreased while KREMEN1 levels were increased (1.5-fold, P = 0.005). The CSNK1E, WNT9A, GSK3A, RARA1, LRP1 and MDM2 gene expression levels in the L cybrids were similar to H cybrids (0.86-fold, P = 0.19; 1.32-fold, P = 0.13; 0.89-fold, P = 0.19, 0.95-fold, P = 0.6; 1.2-fold, P = 0.7; and 0.97-fold, P = 0.77, respectively).
![]()
Table 4: Genes Expression Levels in L Cybrids versus H Cybrids Measured by Q-PCR.
View Table 4
The H and L cybrids also showed differential expression of diabetes-related genes. The diabetes-related gene, ADAMTS9, had elevated expression levels (1.6-fold, P = 0.008) in the L cybrids. The VEGFA and RPS6KA4 gene expression levels were lower in the L cybrids compare to H cybrids (0.79-fold, P = 0.02 and 0.5-fold, P = 0.003, respectively).
Discussion
Different racial/ethnic populations have different susceptibilities to develop specific diseases (Table 5) [6,9]. For example, in populations twenty years or older, the age-adjusted percentage of individuals with diabetes varies: non-Hispanic blacks (13.2%), Hispanic/Latinos (12.8%), Asian-Americans (9.0%) and non-Hispanic whites (7.6%) (http://www.cdc.gov/diabetes/pubs/statsreport14/national-diabetes-report-web.pdf). Approximately, 11% of all African-Americans 20 years of age or older have diabetes and 25% of African-Americans have diabetes by ages 65-74 (http://www.diabetes.niddk.nih.gov/dm/pubs/statistics). African-Americans are twice more likely to have diabetes-related vision loss. In the Canary Islands, there is an ethnic susceptibility to diabetes due directly to maternal mtDNA [28]. In this population, the likelihood of inheritance is higher from an affected mother rather than an affected father [29]. Epidemiological studies illustrate that racial/ethnic disparities can also be found in eye diseases (Table 6) [30-32]. The causes for different susceptibilities to disease are not known, however, there is mounting evidence that mtDNA haplogroups, which define different racial populations, may be a contributing factor [16,33].
![]()
Table 5: Systemic Diseases with Ethnic/Racial Disparities.
View Table 5
![]()
Table 6: Eye Diseases with Ethnic/Racial Disparities.
View Table 6
The DKK and SFRP families of molecules play critical roles in inhibition of the Wnt/β-catenin pathway. By Q-PCR analyses, the African-origin L cybrids showed decreased gene expression levels for both DKK3 and SFRP1, which could lead to increased transcription for downstream genes. DKK3, previously called RIG (Regulated in Glioma), has been associated with tumorigenesis [34-36] and its levels were decreased in malignant glioblastoma cells compared to low-grade astrocytoma and normal brain cells [37]. Low levels of DKK3 and SFRP1 have been reported in high-grade endometrial endometrioid adenocarcinoma and lower progression-free survival [38]. In addition, aberrant methylation status of DKK3 promoter has been associated with human breast cancer [39] and correlated with increasing age in male subjects [40]. DKK3 also plays a role in differentiation of pluripotent stem cells into smooth muscle cells and dopaminergic neurons [41,42] and co-localizes to amyloid-β proteins within senile plaques of Alzheimer’s brains [43]. The differential expressions of DKK3 and SFRP1 in cybrids containing African-origin mtDNA need to be studied further because there is poorer prognosis of various cancers in this population [44-46].
The altered regulation of the WNT/β-catenin pathway also plays a role in the development of diabetes. Genetic studies show that mutations in the LDL receptor proteins (e.g., LRP5 and LRP6) are associated with increased risks of type 1 diabetes, obesity, metabolic syndrome and coronary artery disease [47-50]. In addition, the polymorphisms within the TCF7L2 and WNT5B genes are associated with increased risk of type 2 diabetes [51-54]. These studies demonstrate the Wnt/β-catenin pathway is critical to pre-diabetic and diabetic conditions in different individuals. Therefore, the different expression of Wnt regulatory molecules in the African-origin L cybrids versus the European-origin H cybrids might alter significantly the Wnt/β-catenin signaling and metabolic conditions within the cells and could greatly affect the cellular homeostasis. In African-origin cybrids, the decreased gene expression for the inhibitors DKK3 and SFRP1 suggests that the Wnt/β-catenin signaling pathway would be in the “ON” state, allowing β-catenin to localize into the nucleus to drive transcription and leading to the downstream effects, compared to the European-origin cybrids that would have higher levels of these Wnt inhibitors.
In addition, there are Wnt/β-catenin independent pathways, the Wnt-planar-cell polarity (Wnt-PCP pathway) and the Wnt-calcium pathway (Wnt-Ca2+ pathway), both of which can be regulated by the levels of SFRP1. With decreased expression SFRP1 in cells with the African-origin mtDNA, these pathways would also be affected, possibly contributing to the development of diabetes, cancers and bone diseases [55-57]. The modulation of Wnt pathway expression may play a role in the significantly altered transcription for nuclear genes related to the complement, innate immunity, inflammation, and apoptosis pathways reported in L cybrids compared to H cybrids [16]. Further investigations are needed to understand the influence of the mtDNA on the Wnt/β-catenin independent pathways.
The DKK proteins interact with KREMEN transmembrane receptors to regulate Wnt signaling. By both the GeneChip array and Q-PCR, the African-origin L cybrids had increased expression levels for KREMEN1 compared to the European-origin H cybrids. While the DKK1 protein and lipoprotein receptor-related protein 6 (LRP6) have been studied extensively in other systems, in our GeneChip analyses, the African-origin L cybrids showed similar expression levels for DKK1 (-1.4-fold) and LRP6 (-1.2-fold) genes compared to the European-origin H cybrids, suggesting their transcription levels may not be influenced by the mtDNA variants.
The H and L cybrids also showed differential expression of diabetes-related genes. Our cybrid model shows that the mtDNA variants within the African-origin mtDNA can upregulate expression levels of ADAMTS9, which is associated with insulin resistance and Type 2 diabetes [58]. Studies of European-American and African-American populations show increased association of ADAMTS9 with diabetes and different risk allele burdens between the racial/ethnic groups [27]. The RPS6KA4 and VEGFA gene expression levels were lower in the L cybrids compare to H cybrids. RPS6KA4, which represents the 90kDa polypeptide 4 of the ribosomal S6 kinases (RSK), a family of Ser/Thr kinases, is involved with cell proliferation, maturation, survival, migration, and invasion [59]. Expression levels of RSK can regulate insulin signaling and glucose metabolism [60], key events in maintaining metabolic balance within cells. With lower expression of RPS6KA4 in cells with the African-origin mtDNA, there may be diminished cell proliferation and survival, leading to cell death which is characteristics of diabetic tissues.
The differences between the European-origin cybrids and African-origin cybrids could be due to the numerous changes in amino acids that result from the non-synonymous SNPs found in L haplogroups versus H haplogroups (Figure 1). Comparisons of SNP changes that have occurred over time have shown that in mtDNA coding regions, the ratios of synonymous to non-synonymous SNPs are variable depending upon an African, Asian or European origin [61]. For example, the European JT cluster has a high degree of non-synonymous variability in the MT-ND2 gene compared to the European H or African L haplogroups, while the HV cluster has large non-synonymous variability in the MT-ND5 gene. The African mtDNA lineages are highly conserved in MT-ATP6 and MT-ATP8. These SNP patterns within the haplogroup clusters may play a role in either high risk or protection in some complex diseases [28,62-65]. In any case, in the present study the African-origin L cybrids showed remarkably different expression levels in the DKK3, SFRP1 and KREMEN1 of the Wnt signaling and the diabetes-related RPS6KA4, ADAMTS9 and VEGFA genes compared to the European-origin H cybrids, which may contribute to racial differences in diseases and responses to medications.
Summary
Based upon our data, along with others, we hypothesize that mtDNA haplogroups representing populations of different geographic origins (African versus European) play a role in racial/ethnic susceptibilities to diseases. Our cybrid results show that a person’s mtDNA haplogroup background can act as a “modifier” for the Wnt/β-catenin and diabetes-related genes, and this may influence the cellular homeostasis. Recognizing that the mtDNA haplogroups may contribute to the development and severity of diseases is an important step toward understanding complex diseases. Future work with the cybrid model is needed to identify the retrograde signaling between the mitochondria and nucleus mediating differential transcription of the important Wnt/β-catenin signaling pathway.
Acknowledgements
We wish to thank the subjects who participated in this study.
Ethical Standards
All studies have been approved by the appropriate ethics committee and performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. All individuals involved in this study gave their informed consent prior to their inclusion in this study.
Supported By
Discovery Eye Foundation, Guenther Foundation, Beckman Initiative for Macular Research, Polly and Michael Smith Foundation, Max Factor Family Foundation, Arnold and Mabel Beckman Foundation, Iris and B. Gerald Cantor Foundation.
References
-
Wallace DC (1992) Diseases of the mitochondrial DNA. Annu Rev Biochem 61: 1175-1212.
-
Wallace DC (1994) Mitochondrial DNA mutations in diseases of energy metabolism. J Bioenerg Biomembr 26: 241-250.
-
McFarland R, Turnbull DM (2009) Batteries not included: diagnosis and management of mitochondrial disease. J Intern Med 265: 210-228.
-
Lee C, Yen K, Cohen P (2013) Humanin: a harbinger of mitochondrial-derived peptides? Trends Endocrinol Metab 24: 222-228.
-
Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, et al. (2015) The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 21: 443-454.
-
Hatzfeld JJ, LaVeist TA, Gaston-Johansson FG (2012) Racial/ethnic disparities in the prevalence of selected chronic diseases among US Air Force members, 2008. Prev Chronic Dis 9: E112.
-
Mensah GA, Mokdad AH, Ford ES, Greenlund KJ, Croft JB (2005) State of disparities in cardiovascular health in the United States. Circulation 111: 1233-1241.
-
Kurian AK, Cardarelli KM (2007) Racial and ethnic differences in cardiovascular disease risk factors: a systematic review. Ethn Dis 17: 143-152.
-
Peek ME, Cargill A, Huang ES (2007) Diabetes health disparities: a systematic review of health care interventions. Med Care Res Rev 64: 101S-56S.
-
Feldman CH, Hiraki LT, Liu J, Fischer MA, Solomon DH, et al. (2013) Epidemiology and sociodemographics of systemic lupus erythematosus and lupus nephritis among US adults with Medicaid coverage, 2000-2004. Arthritis Rheum 65: 753-763.
-
Hiraki LT, Feldman CH, Liu J, Alarcon GS, Fischer MA, et al. (2012) Prevalence, incidence, and demographics of systemic lupus erythematosus and lupus nephritis from 2000 to 2004 among children in the US Medicaid beneficiary population. Arthritis Rheum 64: 2669-2676.
-
Manly JJ, Mayeux R (2004) Ethnic differences in dementia and Alzheimer’s disease. In: Anderson N BR, Cohen B, Critical Perspectives on Racial and Ethnic Differences in Health in Late Life. National Academies Press, Washington DC.
-
Dilworth-Anderson P, Hendrie HC, Manly JJ, Khachaturian AS, Fazio S; Social, et al. (2008) Diagnosis and assessment of Alzheimer's disease in diverse populations. Alzheimers Dement 4: 305-309.
-
Potter GG, Plassman BL, Burke JR, Kabeto MU, Langa KM, et al. (2009) Cognitive performance and informant reports in the diagnosis of cognitive impairment and dementia in African Americans and whites. Alzheimers Dement 5: 445-453.
-
Wiley KL, Treadwell E, Manigaba K, Word B, Lyn-Cook BD (2013) Ethnic differences in DNA methyltransferases expression in patients with systemic lupus erythematosus. J Clin Immunol 33: 342-348.
-
Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, et al. (2014) Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: Implications for population susceptibility to diseases. Biochim Biophys Acta 1842:208-219.
-
Gomez-Duran A, Pacheu-Grau D, Martinez-Romero I, Lopez-Gallardo E, Lopez-Perez MJ, et al. (2012) Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of Leber's hereditary optic neuropathy. Biochim Biophys Acta 1822: 1216-1222.
-
Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, et al. (2014) Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum Mol Genet 23: 3537-3551.
-
Kenney MC, Chwa M, Atilano SR, Pavlis JM, Falatoonzadeh P, et al. (2013) Mitochondrial DNA variants mediate energy production and expression levels for CFH, C3 and EFEMP1 genes: implications for age-related macular degeneration. PLoS ONE 8: e54339.
-
Malik D, Hsu T, Falatoonzadeh P, Cáceres-del-Carpio J, Tarek M, et al. (2014) Human retinal transmitochondrial cybrids with J or H mtDNA haplogroups respond differently to ultraviolet radiation: implications for retinal diseases. PLoS One 9: e99003.
-
Miller JR (2002) The Wnts. Genome Biol 3: REVIEWS3001.
-
Jin T, Liu L (2008) The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol 22: 2383-2392.
-
Purro SA, Galli S, Salinas PC (2014) Dysfunction of Wnt signaling and synaptic disassembly in neurodegenerative diseases. J Mol Cell Biol 6: 75-80.
-
Miceli MV, Jazwinski SM (2005) Nuclear gene expression changes due to mitochondrial dysfunction in ARPE-19 cells: implications for age-related macular degeneration. Invest Ophthalmol Vis Sci 46: 1765-1773.
-
Chomyn A. (1996) Platelet-mediated transformation of human mitochondrial DNA-less cells. Rubin. B, editor. Salt Lake City, UT: Academic Press, Inc.
-
Udar N, Atilano SR, Memarzadeh M, Boyer DS, Chwa M, et al. (2009) Mitochondrial DNA haplogroups associated with age-related macular degeneration. Invest Ophthalmol Vis Sci 50: 2966-2974.
-
Keaton JM, Cooke Bailey JN, Palmer ND, Freedman BI, Langefeld CD, et al. (2014) A comparison of type 2 diabetes risk allele load between African Americans and European Americans. Hum Genet 133: 1487-1495.
-
González AM, Maceira BM, Pérez E, Cabrera VM, López AJ, et al. (2012) Genetics, environment, and diabetes-related end-stage renal disease in the Canary Islands. Genet Test Mol Biomarkers 16: 859-864.
-
Boronat M, Chirino R, Varillas VF, Saavedra P, Marrero D, et al. (2005) Prevalence of the metabolic syndrome in the island of Gran Canaria: comparison of three major diagnostic proposals. Diabetic medicine 22: 1751-1756.
-
Bressler SB, Muñoz B, Solomon SD, West SK; Salisbury Eye Evaluation (SEE) Study Team (2008) Racial differences in the prevalence of age-related macular degeneration: the Salisbury Eye Evaluation (SEE) Project. Arch Ophthalmol 126: 241-245.
-
Siegfried CJ, Shui YB, Holekamp NM, Bai F, Beebe DC (2011) Racial differences in ocular oxidative metabolism: implications for ocular disease. Arch Ophthalmol 129: 849-854.
-
Emanuele N, Sacks J, Klein R, Reda D, Anderson R, et al. (2005) Ethnicity, race, and baseline retinopathy correlates in the veterans affairs diabetes trial. Diabetes Care 28: 1954-1958.
-
Sinxadi PZ, Dave JA, Samuels DC, Heckmann JM, Maartens G, et al. (2013) Mitochondrial genomics and antiretroviral therapy-associated metabolic complications in HIV-infected Black South Africans: a pilot study. AIDS Research and Human Retroviruses 29: 1031-1039.
-
Tsuji T, Miyazaki M, Sakaguchi M, Inoue Y, Namba M (2000) A REIC gene shows down-regulation in human immortalized cells and human tumor-derived cell lines. Biochem Biophys Res Commun 268: 20-24.
-
Nozaki I, Tsuji T, Iijima O, Ohmura Y, Andou A, et al. (2001) Reduced expression of REIC/Dkk-3 gene in non-small cell lung cancer. Int J Oncol 19: 117-121.
-
Ryu SW, Kim JH, Kim MK, Lee YJ, Park JS, et al. (2013) Reduced expression of DKK3 is associated with adverse clinical outcomes of uterine cervical squamous cell carcinoma. Int J Gynecol Cancer 23: 134-140.
-
Ligon AH, Pershouse MA, Jasser SA, Yung WK, Steck PA (1997) Identification of a novel gene product, RIG, that is down-regulated in human glioblastoma. Oncogene 14: 1075-1081.
-
Eskander RN, Ali S, Dellinger T, Lankes HA, Randall LM, et al. (2016) Expression Patterns of the Wnt Pathway Inhibitors Dickkopf3 and Secreted Frizzled-Related Proteins 1 and 4 in Endometrial Endometrioid Adenocarcinoma: An NRG Oncology/Gynecologic Oncology Group Study. Int J Gynecol Cancer 26: 125-132.
-
Veeck J, Bektas N, Hartmann A, Kristiansen G, Heindrichs U, et al. (2008) Wnt signalling in human breast cancer: expression of the putative Wnt inhibitor Dickkopf-3 (DKK3) is frequently suppressed by promoter hypermethylation in mammary tumours. Breast Cancer Res 10: R82.
-
Samaei NM, Yazdani Y, Alizadeh-Navaei R, Azadeh H, Farazmandfar T1 (2014) Promoter methylation analyses of WNT/β-catenin pathway regulators and its association with expression of DNMT1 enzyme in colorectal cancer. J Biomed Sci 21: 73.
-
Wang X, Karamariti E, Simpson R, Wang W, Xu Q (2015) Dickkopf Homolog 3 Induces Stem Cell Differentiation into Smooth Muscle Lineage via ATF6 Signalling. J Biol Chem 290: 19844-19852.
-
Fukusumi Y, Meier F, Götz S, Matheus F, Irmler M, et al. (2015) Dickkopf 3 Promotes the Differentiation of a Rostrolateral Midbrain Dopaminergic Neuronal Subset In Vivo and from Pluripotent Stem Cells In Vitro in the Mouse. J Neurosci 35: 13385-13401.
-
Bruggink KA, Kuiperij HB, Gloerich J, Otte-Höller I, Rozemuller AJ, et al. (2015) Dickkopf-related protein 3 is a potential Aβ-associated protein in Alzheimer's Disease. J Neurochem 134: 1152-1162.
-
DeSantis C, Ma J, Bryan L, Jemal A (2014) Breast cancer statistics, 2013. CA Cancer J Clin 64: 52-62.
-
Khawja SN, Mohammed S, Silberfein EJ, Musher BL, Fisher WE, et al. (2015) Pancreatic cancer disparities in African Americans. Pancreas 44: 522-527.
-
Brewster AM, Chavez-MacGregor M, Brown P (2014) Epidemiology, biology, and treatment of triple-negative breast cancer in women of African ancestry. Lancet Oncol 15: e625-634.
-
Twells RC, Mein CA, Payne F, Veijola R, Gilbey M, et al. (2003) Linkage and association mapping of the LRP5 locus on chromosome 11q13 in type 1 diabetes. Hum Genet 113: 99-105.
-
Twells RC, Mein CA, Phillips MS, Hess JF, Veijola R, et al. (2003) Haplotype structure, LD blocks, and uneven recombination within the LRP5 gene. Genome Res 13: 845-855.
-
Mani A, Radhakrishnan J, Wang H, Mani A, Mani MA, et al. (2007) LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science 315: 1278-1282.
-
Lim HW, Lee JE, Shin SJ, Lee YE, Oh SH, et al. (2002) Identification of differentially expressed mRNA during pancreas regeneration of rat by mRNA differential display. Biochem Biophys Res Commun 299: 806-812.
-
Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, et al. (2007) Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest 117: 2155-2163.
-
Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, et al. (2006) TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 355: 241-250.
-
Weedon MN (2007) The importance of TCF7L2. Diabet Med 24: 1062-1066.
-
Kanazawa A, Tsukada S, Sekine A, Tsunoda T, Takahashi A, et al. (2004) Association of the gene encoding wingless-type mammary tumor virus integration-site family member 5B (WNT5B) with type 2 diabetes. Am J Hum Genet 75: 832-843.
-
Fuster JJ, Zuriaga MA, Ngo DT, Farb MG, Aprahamian T, et al. (2015) Noncanonical Wnt signaling promotes obesity-induced adipose tissue inflammation and metabolic dysfunction independent of adipose tissue expansion. Diabetes 64: 1235-1248.
-
Wang Y, Li YP, Paulson C, Shao JZ, Zhang X, et al. (2014) Wnt and the Wnt signaling pathway in bone development and disease. Front Biosci (Landmark Ed) 19: 379-407.
-
Zheng L, Sun D, Fan W, Zhang Z, Li Q, et al. (2015) Diagnostic value of SFRP1 as a favorable predictive and prognostic biomarker in patients with prostate cancer. PLoS One 10: e0118276.
-
Trombetta M, Bonetti S, Boselli ML, Miccoli R, Trabetti E, et al. (2013) PPARG2 Pro12Ala and ADAMTS9 rs4607103 as "insulin resistance loci" and "insulin secretion loci" in Italian individuals. The GENFIEV study and the Verona Newly Diagnosed Type 2 Diabetes Study (VNDS) 4. Acta Diabetol 50: 401-408.
-
Duan RB, Zhang L, Chen DF, Yang F, Yang JS, et al. (2014) Two p90 ribosomal S6 kinase isoforms are involved in the regulation of mitotic and meiotic arrest in Artemia. J Biol Chem 289: 16006-16015.
-
Smadja-Lamere N, Shum M, Deleris P, Roux PP, Abe J, et al. (2013) Insulin activates RSK (p90 ribosomal S6 kinase) to trigger a new negative feedback loop that regulates insulin signaling for glucose metabolism. J Biol Chem 288: 31165-31176.
-
Moilanen JS, Finnila S, Majamaa K (2003) Lineage-specific selection in human mtDNA: lack of polymorphisms in a segment of MTND5 gene in haplogroup J. Mol Biol Evol 20: 2132-2142.
-
Torroni A, Petrozzi M, D'Urbano L, Sellitto D, Zeviani M, et al. (1997) Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am J Hum Genet 60: 1107-1121.
-
Wallace DC, Brown MD, Lott MT (1999) Mitochondrial DNA variation in human evolution and disease. Gene 238: 211-230.
-
Reynier P, Penisson-Besnier I, Moreau C, Savagner F, Vielle B, et al. (1999) mtDNA haplogroup J: a contributing factor of optic neuritis. Eur J Hum Genet 7: 404-406.
-
Hudson G, Nalls M, Evans JR, Breen DP, Winder-Rhodes S, et al. (2013) Two-stage association study and meta-analyses of mitochondrial DNA variants in Parkinson disease. Neurology 80: 2042-2048.