International Journal of Cancer and Clinical Research
Hepatic Cancer Stem Cells and Signaling Pathways
Mohammad Khalid Zakaria1,2, Ashraf Ali3, Kaneez Fatima4, Mohd Suhail3, Shilu Mathew5, Saleh Alkarim6, Esam Azhar7,8 and Ishtiaq Qadri3*
1National Brain Research Centre, Haryana, India
2Department of Biochemistry, All India Institute of Medical Sciences, India
3Department of Medical Biotechnology, King Fahd Medical Research Center, King Abdul Aziz University, Saudi Arabia
4IQ Institute of Infection and Immunity, Pakistan
5Center of Excellence in Genomic Medicine Research, King Abdul Aziz University, Saudi Arabia
6Department of Biological Sciences, King Abdul Aziz University, Saudi Arabia
7Special Infectious Agents Unit-Biosafety Level 3, King Fahd Medical Research Center, King Abdul Aziz University, Kingdom of Saudi Arabia
8Medical Laboratory Technology Department, Faculty of Applied Medical Sciences, King Abdul Aziz University, Kingdom of Saudi Arabia
*Corresponding author:
Ishtiaq Qadri, Department of Medical Biotechnology, King Fahd Medical Research Center, King Abdul Aziz University, PO Box 80216, Jeddah 21589, Saudi Arabia, Tel: +966 640 1000, Fax: +966 6952 5321, E-mail: ishtiaq80262@yahoo.com
Int J Cancer Clin Res, IJCCR-2-017, (Volume 2, Issue 2), Review Article; ISSN: 2378-3419
Received: March 12, 2015 | Accepted: April 28, 2015 | Published: April 30, 2015
Citation: Zakaria MK, Ali A, Fatima K, Suhail M, Mathew S, et al. (2015) Hepatic Cancer Stem Cells and Signaling Pathways. Int J Cancer Clin Res 2:017. 10.23937/2378-3419/2/2/1017
Copyright: © 2015 Zakaria MK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Loss of hepatocytes due to infection, inflammation or partial hepectomy simulates a response which helps in the liver restoration. This maintains homeostasis and keeps a check on the usual wear and tear in the liver. Cancer stem cells (CSCs) were demonstrated to be associated with myeloid leukemia. However, with recent advancements in the approaches and techniques, CSCs are also present within a wide variety of solid tumors and malignancies of epithelial origin. Identification of CSCs has been lately possible by characterization of specific surface markers and signaling events. Hepatocellular carcinoma (HCC) and Cholangiocarcinoma (CC) constitute primary liver cancer. Deaths caused by HCC are much higher than the occurrence possibly due to its asymptomatic nature. The symptoms are manifested at a later stage by the time tumor has metastasized. CSC population, associated with HCC, is majorly responsible for chemo resistance and metastasis. Identifying CSC specific genes, cell surface markers and signaling pathways could help in the development of novel therapeutic approaches. Moreover, the role of micro RNAs (miRNAs) has been shown to be associated with the self-renewal of liver CSCs. This review deals with the understanding of liver CSCs, miRNAs and the signaling pathways involved.
Keywords
Cancer stem cells, Hepatocellular carcinoma, Cholangiocarcinoma, miRNA, Signaling pathways
Introduction
Neoplastic cells are heterogeneous in nature [1]. Such contrasting nature has been described due to the accumulation of epigenetic [2] and genetic [3] changes. Variability in the tumor microenvironment might activate specific signaling cascades within the tumor cells. This possibly contribute towards heterogeneity [4]. Additionally, such heterogeneity could also be contributed by cells with stem cell like properties and are called as cancer stem cells (CSCs; [5]). Tumor propagation, as a result of CSCs, has been experimentally proved by characterization of cells with stem cell like properties (self-renewal, positive for stem cell surface markers) [6,7]. Various tumorigenic and clonogenic assays have established the self-renewal properties of these CSCs. CSCs assist in the development of tumor by both symmetrical and asymmetrical division, chemo-resistance, radio-resistance and metastasis [7]. It can be assumed that the CSCs are the major players of tumorigenesis and are important therapeutic targets as they are implicated in a wide variety of cancers including hepatocellular carcinoma (HCC). HCC is the third most leading cause of cancer deaths. However, on the level of occurrence, it is sixth. This is possibly due to its asymptomatic nature [8]. HCC constitutes around 75% of the primary liver cancer cases [9]. Morphological heterogeneity exists in the case of HCC and intra-hepatic cholangiocarcinoma (ICC;[10]. Self-renewal capability of CSCs has been well established through xenotransplantation of HCC specimens. A variety of CSCs surface markers have been identified. These include EpCAM, CD-44,40,24,13,133, OV6, aldehyde dehydrogenase activity etc. Some of these also contribute to characteristics such as chemoresistance and invasive potential in HCC[11]. Several reports have confirmed the role of micro RNAs (miRNAs) in liver CSCs. Some of these miRNAs are the major regulators of CSC maintenance and self-renewal. One of the possible mechanisms that keep the CSCs in an undifferentiated state could be by means of miRNAs. Furthermore, down-regulation of these miRNAs has shown to induce differentiation of CSCs. This suggests their critical role in CSCs regulation. Chemo resistance, acquired by these CSCs, is also believed to be mediated by various miRNAs [12]. De-regulation of signaling pathway is another factor responsible for transforming normal liver stem cells into CSCs. Some of the pathways activated in liver CSCs are Hedgehog, Wnt/β-catenin, IL-6/STAT3, Oncostatin M (OSM), bone morphogenic proteins (BMPs), BMI I, Notch, TGF-β [11]. These signaling events contribute to the survival and propagation of liver CSCs.
Hepatic Stem Cell Pool
Hepatocytes
Hepatocytes are non-dividing during the majority of their cell cycle. Upon cell loss, due to damage, infection and inflammation, hepatocytes divide in order to restore liver mass. In rodents, liver can restore up to 2/3rd of the mass after partial hepectomy [13,14]. Hepatocytes divide actively in the periportal region of the liver in response to the regenerative stimuli. This hepatocyte division, from the periphery, reaches the central region. Studies have successfully demonstrated the infinite capacity of hepatocyte division within the animal models [15]. In rats, there is a direct relation between HCC cells and hepatocytes. A group showed that a significant percentage of regenerative hepatocytes were produced directly from mature hepatocytes. When HCC was induced chemically by diethyl nitrosamine (DEN), approximately similar proportion of HCC originated from mature hepatocytes indicating a close association of HCC and mature hepatocytes [16].
Several studies have also shown the self-renewal potential of hepatocytes in animal systems. F344 rats, with partial hepectomy, were treated with retrorsine to inhibit hepatocyte replication. Transfer of hepatocytes, from external source, could also replace the liver mass completely [17]. Contrastingly, when rats were treated with retrorsine before partial hepectomy, complete liver regeneration was observed. This was shown to be by small hepatocyte-like progenitor cells (SHPCs) [18]. It also indicates that when replication is inhibited, differentiation takes over.
Chronic hepatitis infection (CHC) leads to the killing of hepatocytes. In order to maintain liver mass proliferation of hepatocytes take place [19]. Such increase in repeated proliferation cycle could also result in the progression of HCC. A group reported that an increased proliferation in hepatitis C subjects ultimately inflicted histological damage to the hepatocytes. This resulted in cirrhosis possibly due senescence [20]. A close association between senescent mature hepatocytes and oval cells has been observed.
Oval cells
These have a high nucleus to cytoplasm ratio and are identified by their characteristic ovoid nucleus. Oval cells possess the capacity to pass through the bile canaliculi [21]. Oval cells can differentiate into either cholangiocytes or hepatocytes [22,23]. Activation studies on oval cells have been conducted in rodents where hepatocyte replication was suppressed by a carcinogen following proliferative stimuli [24]. Also, in mice where hepatocyte regeneration was repressed by oxidative stress, activation of oval cells was observed [25]. Yasui and coworkers showed the generation of albumin secreting hepatocytes from oval cells [26]. In the case of viral hepatitis, fatty liver disease and necrosis, mature hepatocytes are not able to regenerate due to the exhaustion or inhibition. Under such conditions stem cell compartments, bearing resemblance to oval cells, have been found. Moreover, stimulation of such progenitor cells corroborates with the degree of damage [27,28]. These progenitor cells could serve as an indicative marker of various liver based diseases. Similarly, in the case of hepatitis, such progenitor cell induction indicates the level of inflammation [29]. There are various markers for the identification of oval cell population such as - AFP, NCAM1, Chro A, OV6 etc. It is also considered that the oval cells are produced by Bone Marrow cells (BMCs), since these cells express some common surface markers.
Bone marrow cells
Hepatocytes and oval cells are derived from BMCs as a regenerative response following liver damage [30]. Sell and coworkers for the first time demonstrated BMCs as the source of oval cells post periportal necrosis [31]. Another transplantation approach showed that in the absence of injury a small proportion of hepatocytes were derived from BMCs. This indicated that the hepatocyte maintenance was being governed by BMCs [32]. Evidence of liver stem cells, origination from BMCs, came from a study by Theise and coworkers. They proved that the patients receiving bone marrow or liver transplant, from that of opposite gender, possessed various proportions of donor derived cells [32,33].
With respect to the liver diseases, the exact role of BMC is still not fully understood. The fact that damaged hepatocytes could alter the lineage commitment of hematopoietic stem cells to hepatocytes could not be avoided. However, most of the current reports highlight only few scenarios where this type of lineage commitment is possible [34].
Linking Cancer Stem Cells and Liver Cancer
Clinically both HCC and CC may coexist. This gives an insight into a close association of stem or progenitor cell in neoplastic tumours. Several reports have already described the influence of liver stem cells in HCC development. Activity of stem and progenitor cells has been closely monitored and is linked with inflammation and fibrosis during hepatitis. Direct role of hepatocytes have been described in liver carcinogenesis [35,36]. Transforming ability of oval cells was demonstrated in nude mice where they developed into tumor mass [37]. However, the role of BMCs in hepatocarcinogenesis is still unclear and needs further study.
Prolonged sustenance of self-renewal signal have the potential of initiating neoplastic transformation [38]. Wnt/β-Catenin and BMI-I are the 2 major signaling cascades known to induce carcinogenesis in liver stem and progenitor cells. In chronic hepatitis and liver injury continuous self-renewal of hepatocytes, through stem/progenitor cells, is known to initiate tumorigenesis [39]. It is believed that the tumor mass comprises of undifferentiated CSCs and partially differentiated non-tumorous cancer cells originating from CSCs. However. only CSCs has the stem cell properties important for survival and are implicated in tumor growth [40].
Identification of Liver Cancer Stem cells
A distinct criterion is set to identify and distinguish CSCs from the usual liver cell population. This has been possible with the help of characteristic markers expressed by CSCs. Hepatic progenitor cells and CSCs express a variety of surface specific molecules that have been characterized. It has also been shown that these markers are implicated in maintaining CSCs characteristics. For instance a variant of CD44 supports and protects CSCs against oxidative stress [41]. Moreover, CD13 confers resistance against genotoxic substances [42]. EpCAM up-regulates Wnt signaling in ES and cancer cells [43,44]. CD133 helps in the maintenance of CD133+ liver CSCs by activating neurotensin/IL-8/CXCL1 signaling [45]. In addition, a CD44 variant maintains oxidative homeostasis by stabilizing xCT, hence protecting liver CSCs from oxidative stress [46]. CD13 also reduces the damage caused by oxidative stress [47]. Thus, the CSC markers could be a good potential target for the eradication of liver CSCs.
Role of miRNAs in Hepatic Cancer Stem Cells
These are non-coding RNAs regulating expression of a variety of genes. Role of miRNA in disease progression has been well established. Reports indicate that the miRNAs are important regulators of cellular protein and helps maintain balance. By down/up-regulation of miRNAs, researchers have established their role and their pathological processes. Expression pattern of miRNAs have been different in liver CSCs and non-CSCs population. miRNA plays a crucial role in CSC self-renewal, resistance and tumor development [12]. miRNA are also being utilized as prognostic markers, since they are stable in the serum of HCC patients.
Liver CSC population has been shown to express some members of miRNA-181 family. This include miRNA-181a,b,c and d. These miRNAs have also shown to be associated with increased tumorigenic potential through proliferative markers such as CDX2, UGT2B7, CYTP3A4 and β-catenin related genes. Additionally, 181s family miRNAs have a role in the maintenance of stemness by regulating COX 2 and GATA 6. miRNA 181 family is directly regulated by Wnt/β-catenin pathway since the down-regulation of Wnt/β-Catenin results in its suppression. This suggests that the repression of miRNA 181 could help prevent HCC [48,49]. Level of miRNA 145 is lower in CSCs as compared to normal ones. miRNA 145 expression in HCC mice model results in the suppression of tumor development. Increased expression of this miRNA led to the decrease in Oct 4 levels suggesting that the anti-tumor effects are mediated by alteration on stem cell marker [50].
miRNA 150 is selectively expressed by liver CSCs which are CD133+. Increased expression of this miRNA results in the suppression of CSCs. Also, increased expression of miRNA 150 decreases cyclin D1 and Bcl-2 ultimately reducing cell survival [51]. Meng and coworkers demonstrated the selective expression of miRNA let-7 in Oct4+ and CD133+ liver CSCs. Let-7 expression was found to be insignificant in normal liver stem cells. Furthermore, upon suppression of let-7, CSCs were sensitized to doxorubicin and sorafenib [52]. IL-6, a direct regulator of let-7a and let-7b, is frequently over expressed in HCC [53]. miRNA 199a-3p has the tendency to decrease the proliferative potential of CD44+ liver CSCs in HCC cell lines. However, presence of this miRNA in HCC patient's sera is still to be reported [54]. A group reported the selective up-regulation of miRNA 130b in CD133 positive liver CSCs extracted from HCC cell lines as well as tissues. miRNA 130b expression enhanced self-renewal potential of these cells. Knocking down miRNA 130b reversed this effect [55]. These reports demonstrate that the miRNAs play an important role in CSCs maintenance.
Signaling Pathways of liver CSCs
BMP
A multipotent foregut endoderm is responsible for giving rise to a vertebrate liver. This tissue is also a source for pancreas, thyroid and lung development [56]. One of the critical developmental regulators is bone morphogenic proteins (BMPs). During liver development, BMPs are produced within the mesoderm adjoining the foregut endoderm. However, the exact sites responsible for BMP production are still not fully studied. BMP belong to TGF-β family and they initiate cell affects by binding to their specific receptors[57]. Ligand specificity is determined by the combination of various receptors. Upon ligand binding, activated BMP receptor (BMPR) phosphorylates SMAD proteins. Furthermore, SMAD proteins dimerises with SMAD 4 and translocate into the nucleus where it regulate transcription of various genes [58]. Previously, BMP pathway has been implicated in colorectal and glioblastoma development. A research group has shown that by exogenously expressing BMP4, CSCs can be differentiated into HCC. Interestingly, endogenous BMP4 helped in CD133 expression in CSCs. SMAD6, a target gene of BMP signaling, corroborated with the expression of CD133 in HCC. The mechanistic study revealed that the differentiation of CSC occurred as a result of Erk1/2 induction by BMP4 [59].
Hedgehog
Hedgehog pathway is highly conserved in humans as well as in drosophila. It has a critical role in maintaining cell fate in adults and during embryonic development [60]. Desert hedgehog (DHH),Sonic hedgehog (SHH) and Indian hedgehog (IHH) ligands seem to exist which bind to membrane based patched (Ptc) receptors [61]. When the ligand is absent, Ptc repress the pathway by binding to another trans membrane protein smoothened (smo; [62]). Hedgehog activation occurs upon binding of ligands to the Ptc, ultimately activating transcription factors Gli 1, 2 and 3. Gli 1 and Gli 2 generally serves as activators, whereas Gli 3 act as a repressor [63]. Within the liver, SHH is the major form and is found in approximately 60% of the HCCs. Reports have validated further that blocking hedgehog pathway could down-regulate Gli based target genes [64]. Also, during neoplastic transformation, proto-oncogene c-Myc expression is increased by smo [65]. Recent reports have established the role of hedgehog signaling in liver CSCs [66].
BMI
Evolutionary conserved group of genes, known as polycomb, constitute BMI I pathway. BMI I has a role in epigenetic modulation of stem cells and helps in the self-renewal process [67]. BMI I over-expression is known to be associated with HCC phenotype [68]. De-regulated BMI I has also been validated in CSC population and is critically involved in the maintenance of liver. The critical role play of BMI I, in liver CSCs propagation, was confirmed by studies on hepatic stem cells with ectopically induced BMI I. BMI I, along with Wnt/β-Catenin pathway, was shown to help in the liver CSCs and stem cell maintenance [69].
Oncostatin M
OSM is produced by CD45+ hematopoietic cells and is a cytokine related to IL-6. OSM is also known to regulate other important liver functions such as ammonia clearance, glycogen/lipid synthesis and detoxification [70]. OSM receptor (OSMR) is composed of OSM specific subunits and gp130 [71]. This leads to the activation of two pathways - STAT3 or Ras [72,73]. OSM causes the differentiation of hepatoblast through the STAT3 signaling cascade [70]. A study shows the differentiation of liver CSCs when OSMR is activated upon OSM binding indicating the importance of OSM signaling in liver CSCs [74].
IL-6/STAT3
Within the liver, macrophages are responsible for the production of IL-6. IL-6, a cytokine, binds to a receptor forming a complex which in turn binds to gp130. Along with IL-6, OSM, LEF and CNTF also share gp130 receptor [75]. Expression profiling of certain HCC tumors have shown the activation of IL-6 signaling pathway. This led to a hypothesis that the development of HCC could be from CSCs with de-regulated IL-6 cascade. This was confirmed by utilizing IL-6 deficient mouse models where marked decrease in HCC was observed [76]. Regeneration of liver is governed by several molecular factors, cytokines and hormones with linkage to their downstream signaling events. STAT3 is the downstream molecule of IL-6 signaling and has a major role in liver regeneration [77,78]. Other cytokines, apart from IL-6, are also known to activate STAT3 and are implicated in liver regeneration [79-81]. IL-10, an anti-inflammatory cytokine, inhibits STAT3 and prevent liver regeneration [82]. Both IL-6 and STAT3 are implicated in maintenance of stem cell self-renewal and also has a likely role in liver CSCs. Moreover, OSM [80] and cardiotrophin1 [83], an IL-6 family cytokines, contribute towards liver proliferative potential. Some reports suggest that there is an inter play between STAT3 of hepatocytes and myeloid cells for liver regeneration. SOCS3 is a negative regulator of STAT3 and is induced by STAT3 itself [84]. In conclusion, it is accepted that STAT3 promotes liver regeneration. However, communication between different liver cells in liver regeneration, via STAT3, needs to be further studied.
Notch/Wnt
Various signaling events seem to exist which regulate stem cell proliferation. Notch signaling is one of the crucial pathways participating actively in the stem cell renewal process [85,86]. Over expression of jagged, a notch ligand, and notch3 has been seen in HCC [87,88]. Notch dependent transformation is linked to the activation of ERK. This leads to the stabilization of Notch transcript and hence active transcription of Hes-1 [89,90]. Contrastingly, Notch can also serve as a tumor suppressor by interacting with other pathways such as Ras/Raf/MEK/ERK and regulating tumor suppressor PTEN [91]. Notch signaling has a crucial role in the liver development and formation of the bile duct. Marked increase in the expression levels of notch genes has been demonstrated in CD133+ liver cancer cells, corroborating with the fact that CSCs have activated Notch Pathway. Wnt signaling pathway also regulates stem and progenitor cells [92]. Wnt signaling stabilizes β-catenin, which activates a family of transcription factor TCF [93]. Receptor of Wnt is known as frizzled (Fz). Activation of Fz with its co-receptor LRP-5/6 leads to the activation of disheveled. This event dissociates tetrameric GSK3β/β-catenin/APC/Axin complex and reduces phosphorylation of β-catenin, mediating its nuclear entry and activating transcription of target genes with the help of TCF. Molecular signaling between Wnt, HH, BMP and Notch is known to regulate stem cell differentiation or self-renewal [94]. More than 80% of HCCs are found to over express FZD-7 [95]. Also, around 20-40% of HCCs showed nuclear and cytoplasmic accumulation of β-catenin [96]. Even though there is a marked increase in β-catenin, its target genes are not affected. This correlates with the fact that the transcriptional activation of Wnt target genes are also regulated by other signaling molecules [97]. In about 25% of HCCs β-catenin and Axin1 mutations have been demonstrated [98,99]. In EpCAM + liver CSCs, over expression of Wnt pathway mediators have been shown [100]. Researchers have demonstrated that the self-renewal potential of stem/progenitor cells could be increased by exogenously expressing mutant β-catenin in mouse hepatic stem or progenitor cells. Also, activation of Wnt/β-catenin pathway has been observed in oval and OV6 + rodent tumor cells. Such activation of Wnt attributes chemo resistance to HCC cells [101]. These findings denote that the Wnt signaling plays an important role in CSC maintenance.
TGF-β
TGF-β signaling has a role in the suppression of foregut cancer as well as in the normal development of the gut endoderm [102]. SMAD signaling has been shown to be important in the proliferation of hepatocytes of the embryonic origin [103]. Activation of SMAD could be initiated by receptors or SMAD binding proteins. These proteins belong to a variety of families such as SUMO ligases, SARA, β-2-Spectrin and Filamin. TGF-β signaling is dependent upon β-2-Spectrin for its activation [104]. β-2-Spectrin presents SMAD3 to the cytoplasmic domain of TGF-β type I receptor complex. This is followed by complex formation with SMAD4 and its subsequent nuclear localization inducing target gene activation [105]. Deficiency of β-2-Spectrin leads to the defective TGF-β pathway. SMAD 2 and 3 heterozygous mice die due to the liver and gastrointestinal defects. This suggests the crucial role of TGF-β pathway in liver development and regeneration. Several reports have shown significant reduction in the levels of TGF-β receptors in liver cancer [106]. However, HCC patients show marked increase of TGF-β in the urine and serum [107]. Also, immuno histochemical studies have confirmed that TGF-β is increased in significant proportion of HCC cases [108]. TGF-β pathway mediates both tumor and anti-tumor effects. In HCC, the anti-proliferative arm of this pathway is lost, thus leading to increased proliferation and survival of tumor cells. TGF-β signaling also caused endothelial to mesenchyme transition (EMT) in neoplastic cells. It is shown by Yuan and coworkers that HCC cells which are Oct4/STAT3 positive and have impaired TGF-β signaling possess the potential in HCC development by acting as CSCs [109].
Other Factors Modulating Cancer Stem Cells
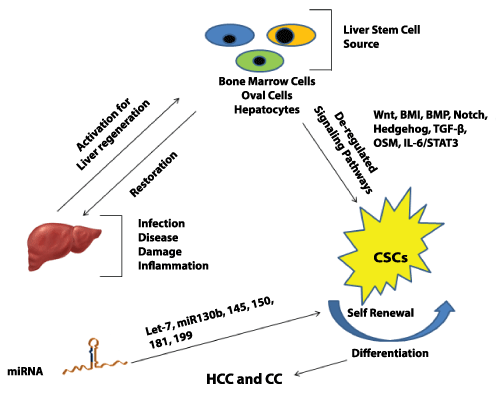
Nobel laureate Yamanaka and his coworkers demonstrated the cellular re-programming of differentiated cells to pluoripotency. This was achieved by selective expression of factors such as Oct3/4, Sox-2, Klf-4, myc etc [110,111]]. Such critical transcriptional molecular factors might also be involved in liver transformation and CSC maintenance. A group showed the existence of hepatic CSCs in c-Myc dependent tumors. This could possibly indicate the role of myc in CSCs and their self-renewal [112]. CD24 and CD133 expressing liver CSCs require Nanog, a transcriptional factor active in ES cells, for self-renewal [113]. P53 loss has been shown to assist in reprogramming by Yamanaka factors [114]. Mutations in TP53 are associated with specific gene signatures linked with stem cell like characteristics [115]. This suggests that the differentiation factors may also enhance HCC progression by maintenance of liver CSC population. Figure 1 shows a summarized view depicting pathways activated in CSCs and miRNAs responsible for their maintenance.
.
Figure 1: Signaling pathways activated in liver cancer stem cell and the miRNAs involved in chemo resistance and self-renewal of these cells
View Figure 1
Conclusion
According to the CSCs hypothesis, tumor comprise of cells heterogeneous in function and phenotype. A small fraction of such cells possess stem cell like features and are responsible for maintenance and development of tumor [116]. In the near future, approaches that selectively target CSCs will be promising. Hence, understanding the biological factors and signals involved in the maintenance and differentiation of CSCs could help develop novel therapeutic strategies to tackle cancer. Currently employed cancer therapies aim to reduce tumor load by inducing cancer cell death. However, tumor recurrence and metastasis are the major hurdles to deal with. CSCs, like normal cells, are often dormant for long durations. Activation signals appear as the environment becomes favorable. Reports have shown that only a small fraction of these CSCs can give rise to tumors that depict the similar phenotype as of the original tumor. Pathways such as Wnt, HH, Notch, IL-6/STAT3, BMP, BMI, OSM, TGF-β seem to have a critical role in the maintenance of such CSC population. Therefore, controlled and selective suppression of these pathways, along with currently employed therapeutic approaches, might prevent tumor recurrence and metastasis and increase the chances of survival. Also, combinatorial approaches inhibiting miRNAs and signaling pathways, implicated in CSCs sustenance, could suppress cancer progression with better outcomes.
Few of the clinical trials, dealing with cancer stem cells and therapy, are ongoing or have been completed at various research centers across the globe. For instance, Ning et al. have successfully devised lung cancer stem cell specific vaccine [117] and their study have been completed till phase 2 clinical trial. Others include vaccine based immunotherapies for HCC and colorectal cancer which are undergoing at the phase 2 level. (Clinical trial number NCT02089919 and NCT02176746 respectively). Phase I-II study with autologous mesenchymal stem cell injection, for cirrhotic patients, have been successfully completed (Clinical trial number NCT00420134). Lastly, the efficacy of bone marrow cells is being estimated in an ongoing trial for treating HBV related cirrhosis (clinical trial number NCT01724697).
References-
Fialkow PJ (1976) Clonal origin of human tumors. Biochim Biophys Acta 458: 283-321.
-
Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 11: 726-734.
-
Nowell PC (1976) The clonal evolution of tumor cell populations. Science 194: 23-28.
-
Mueller MM, Fusenig NE (2004) Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer 4: 839-849.
-
Jordan CT, Guzman ML, Noble M (2006) Cancer stem cells. N Engl J Med 355: 1253-1261.
-
Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, et al. (1994) A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367: 645-648.
-
Magee JA, Piskounova E, Morrison SJ (2012) Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 21: 283-296.
-
Forner A, Llovet JM, Bruix J (2012) Hepatocellular carcinoma. Lancet 379: 1245-1255.
-
Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011) Global cancer statistics. CA Cancer J Clin 61: 69-90.
-
Zhang F, Chen XP, Zhang W, Dong HH, Xiang S, et al. (2008) Combined hepatocellular cholangiocarcinoma originating from hepatic progenitor cells: immunohistochemical and double-fluorescence immunostaining evidence. Histopathology 52: 224-232.
-
Yamashita T, Wang XW (2013) Cancer stem cells in the development of liver cancer. J Clin Invest 123: 1911-1918.
-
Kasinski AL, Slack FJ (2011) Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 11: 849-864.
-
Fausto N (2004) Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology 39: 1477-1487.
-
Michalopoulos GK, DeFrances MC (1997) Liver regeneration. Science 276: 60-66.
-
Overturf K, al-Dhalimy M, Ou CN, Finegold M, Grompe M (1997) Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol 151: 1273-1280.
-
Bralet MP, Pichard V, Ferry N (2002) Demonstration of direct lineage between hepatocytes and hepatocellular carcinoma in diethylnitrosamine-treated rats. Hepatology 36: 623-630.
-
Laconi E, Oren R, Mukhopadhyay DK, Hurston E, Laconi S, et al. (1998) Long-term, near-total liver replacement by transplantation of isolated hepatocytes in rats treated with retrorsine. Am J Pathol 153: 319-329.
-
Avril A, Pichard V, Bralet MP, Ferry N (2004) Mature hepatocytes are the source of small hepatocyte-like progenitor cells in the retrorsine model of liver injury. J Hepatol 41: 737-743.
-
Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, et al. (1996) Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci U S A 93: 4398-4402.
-
Falkowski O, An HJ, Ianus IA, Chiriboga L, Yee H, et al. (2003) Regeneration of hepatocyte 'buds' in cirrhosis from intrabiliary stem cells. J Hepatol 39: 357-364.
-
Marrero JA (2005) Hepatocellular carcinoma. Curr Opin Gastroenterol 21: 308-312.
-
Kim H, Park C, Han KH, Choi J, Kim YB, et al. (2004) Primary liver carcinoma of intermediate (hepatocyte-cholangiocyte) phenotype. J Hepatol 40: 298-304.
-
Robrechts C, De Vos R, Van den Heuvel M, Van Cutsem E, Van Damme B, et al. (1998) Primary liver tumour of intermediate (hepatocyte-bile duct cell) phenotype: a progenitor cell tumour? Liver 18: 288-293.
-
Alison MR, Golding M, Sarraf CE, Edwards RJ, Lalani EN (1996) Liver damage in the rat induces hepatocyte stem cells from biliary epithelial cells. Gastroenterology 110: 1182-1190.
-
Roskams T, Desmet V (1998) Ductular reaction and its diagnostic significance. Semin Diagn Pathol 15: 259-269.
-
Yasui O, Miura N, Terada K, Kawarada Y, Koyama K, et al. (1997) Isolation of oval cells from Long-Evans Cinnamon rats and their transformation into hepatocytes in vivo in the rat liver. Hepatology 25: 329-334.
-
Libbrecht L, Desmet V, Roskams T (2005) Preneoplastic lesions in human hepatocarcinogenesis. Liver Int 25: 16-27.
-
Lowes KN, Brennan BA, Yeoh GC, Olynyk JK (1999) Oval cell numbers in human chronic liver diseases are directly related to disease severity. Am J Pathol 154: 537-541.
-
Libbrecht L, Desmet V, Van Damme B, Roskams T (2000) Deep intralobular extension of human hepatic 'progenitor cells' correlates with parenchymal inflammation in chronic viral hepatitis: can 'progenitor cells' migrate? J Pathol 192: 373-378.
-
Petersen BE, Bowen WC, Patrene KD, Mars WM, Sullivan AK, et al. (1999) Bone marrow as a potential source of hepatic oval cells. Science 284: 1168-1170.
-
Yin L, Lynch D, Sell S (1999) Participation of different cell types in the restitutive response of the rat liver to periportal injury induced by allyl alcohol. J Hepatol 31: 497-507.
-
Theise ND, Nimmakayalu M, Gardner R, Illei PB, Morgan G, et al. (2000) Liver from bone marrow in humans. Hepatology 32: 11-16.
-
Alison MR, Poulsom R, Jeffery R, Dhillon AP, Quaglia A, et al. (2000) Hepatocytes from non-hepatic adult stem cells. Nature 406: 257.
-
Thorgeirsson SS, Grisham JW (2006) Hematopoietic cells as hepatocyte stem cells: a critical review of the evidence. Hepatology 43: 2-8.
-
Tatematsu M, Nagamine Y, Farber E (1983) Redifferentiation as a basis for remodeling of carcinogen-induced hepatocyte nodules to normal appearing liver. Cancer Res 43: 5049-5058.
-
Williams GM, Gebhardt R, Sirma H, Stenbäck F (1993) Non-linearity of neoplastic conversion induced in rat liver by low exposures to diethylnitrosamine. Carcinogenesis 14: 2149-2156.
-
Dumble ML, Croager EJ, Yeoh GC, Quail EA (2002) Generation and characterization of p53 null transformed hepatic progenitor cells: oval cells give rise to hepatocellular carcinoma. Carcinogenesis 23: 435-445.
-
Wicha MS, Liu S, Dontu G (2006) Cancer stem cells: an old idea--a paradigm shift. Cancer Res 66: 1883-1890.
-
Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, et al. (2008) Cancer stem cell markers in common cancers - therapeutic implications. Trends Mol Med 14: 450-460.
-
Zeng G, Apte U, Cieply B, Singh S, Monga SP (2007) siRNA-mediated beta-catenin knockdown in human hepatoma cells results in decreased growth and survival. Neoplasia 9: 951-959.
-
Kuvshinoff BW, Ota DM (2002) Radiofrequency ablation of liver tumors: influence of technique and tumor size. Surgery 132: 605-611.
-
Huang H CY, Chuang CY, Stone L, et al. (2011) Epithelial cell adhesion molecule (EpCAM) complex proteins promote transcription factor-mediated pluripotency reprogramming. J Biol Chem 286: 33520-33532.
-
Tang KH, Ma S, Lee TK, Chan YP, Kwan PS (2012) CD133(+) liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology Md55: 807-820.
-
Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, et al. (2011) CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell 19: 387-400.
-
Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M, et al. (2010) CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest 120: 3326-3339.
-
Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, et al. (2009) Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology 50: 472-480.
-
Ji J, Yamashita T, Wang XW (2011) Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci 1: 4.
-
Jia Y, Liu H, Zhuang Q, Xu S, Yang Z, et al. (2012) Tumorigenicity of cancer stem-like cells derived from hepatocarcinoma is regulated by microRNA-145. Oncol Rep 27: 1865-1872.
-
Zhang J, Luo N, Luo Y, Peng Z, Zhang T, et al. (2012) microRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb. Int J Oncol 40: 747-756.
-
Meng F, Glaser SS, Francis H, DeMorrow S, Han Y, et al. (2012) Functional analysis of microRNAs in human hepatocellular cancer stem cells. J Cell Mol Med 16: 160-173.
-
Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, et al. (2010) Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res 70: 9798-9807.
-
Henry JC, Park JK, Jiang J, Kim JH, Nagorney DM, et al. (2010) miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun 403: 120-125.
-
Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, et al. (2010) miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 7: 694-707.
-
Wells JM, Melton DA (1999) Vertebrate endoderm development. Annu Rev Cell Dev Biol 15: 393-410.
-
Chen D, Ji X, Harris MA, Feng JQ, Karsenty G, et al. (1998) Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J Cell Biol 142: 295-305.
-
Derynck R, Zhang YE (2003) Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425: 577-584.
-
Zhang L, Sun H, Zhao F, Lu P, Ge C, et al. (2012) BMP4 administration induces differentiation of CD133+ hepatic cancer stem cells, blocking their contributions to hepatocellular carcinoma. Cancer Res.72, 4276-4285.
-
McMahon AP, Ingham PW, Tabin CJ (2003) Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol 53: 1-114.
-
Ingham PW, McMahon AP (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15: 3059-3087.
-
Alcedo J, Noll M (1997) Hedgehog and its patched-smoothened receptor complex: a novel signalling mechanism at the cell surface. Biol Chem 378: 583-590.
-
Koebernick K, Pieler T (2002) Gli-type zinc finger proteins as bipotential transducers of Hedgehog signaling. Differentiation 70: 69-76.
-
Sicklick JK, Li YX, Jayaraman A, Kannangai R, Qi Y, et al. (2006) Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis 27: 748-757.
-
Huang S, He J, Zhang X, Bian Y, Yang L, et al. (2006) Activation of the hedgehog pathway in human hepatocellular carcinomas. Carcinogenesis 27: 1334-1340.
-
Oishi N, Wang XW (2011) Novel therapeutic strategies for targeting liver cancer stem cells. Int J Biol Sci 7: 517-535.
-
Park IK, Qian D, Kiel M, Becker MW, Pihalja M, et al. (2003) Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302-305.
-
Sasaki M, Ikeda H, Itatsu K, Yamaguchi J, Sawada S, et al. (2008) The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab Invest 88: 873-882.
-
Chiba T, Seki A, Aoki R, Ichikawa H, Negishi M, et al. (2010) Bmi1 promotes hepatic stem cell expansion and tumorigenicity in both Ink4a/Arf-dependent and -independent manners in mice. Hepatology 52: 1111-1123.
-
Kamiya A, Kinoshita T, Ito Y, Matsui T, Morikawa Y, et al. (1999) Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J 18: 2127-2136.
-
Lindberg RA, Juan TS, Welcher AA, Sun Y, Cupples R, et al. (1998) Cloning and characterization of a specific receptor for mouse oncostatin M. Mol Cell Biol 18: 3357-3367.
-
Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, et al. (1996) Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity 5: 449-460.
-
Hirano T, Nakajima K, Hibi M (1997) Signaling mechanisms through gp130: a model of the cytokine system. Cytokine Growth Factor Rev 8: 241-252.
-
Kinoshita T, Miyajima A (2002) Cytokine regulation of liver development. Biochim Biophys Acta 1592: 303-312.
-
Benigni F, Fantuzzi G, Sacco S, Sironi M, Pozzi P, et al. (1996) Six different cytokines that share GP130 as a receptor subunit, induce serum amyloid A and potentiate the induction of interleukin-6 and the activation of the hypothalamus-pituitary-adrenal axis by interleukin-1. Blood 87: 1851-1854.
-
Mishra L, Banker T, Murray J, Byers S, Thenappan A, et al. (2009) Liver stem cells and hepatocellular carcinoma. Hepatology 49: 318-329.
-
Michalopoulos GK (2007) Liver regeneration. J Cell Physiol 213: 286-300.
-
Taub R (2004) Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol 5: 836-847.
-
Beraza N, Marqués JM, Martínez-Ansó E, Iñiguez M, Prieto J, et al. (2005) Interplay among cardiotrophin-1, prostaglandins, and vascular endothelial growth factor in rat liver regeneration. Hepatology 41: 460-469.
-
Nakamura K, Nonaka H, Saito H, Tanaka M, Miyajima A (2004) Hepatocyte proliferation and tissue remodeling is impaired after liver injury in oncostatin M receptor knockout mice. Hepatology 39: 635-644.
-
Ren X, Hu B, Colletti LM (2010) IL-22 is involved in liver regeneration after hepatectomy. Am J Physiol Gastrointest Liver Physiol 298: G74-80.
-
Yin S, Wang H, Park O, Wei W, Shen J, et al. (2011) Enhanced liver regeneration in IL-10-deficient mice after partial hepatectomy via stimulating inflammatory response and activating hepatocyte STAT3. Am J Pathol 178: 1614-1621.
-
Bustos M, Beraza N, Lasarte JJ, Baixeras E, Alzuguren P, et al. (2003) Protection against liver damage by cardiotrophin-1: a hepatocyte survival factor up-regulated in the regenerating liver in rats. Gastroenterology 125: 192-201.
-
Riehle KJ, Campbell JS, McMahan RS, Johnson MM, Beyer RP, et al. (2008) Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J Exp Med 205: 91-103.
-
Androutsellis-Theotokis A1, Leker RR, Soldner F, Hoeppner DJ, Ravin R, et al. (2006) Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 442: 823-826.
-
Artavanis-Tsakonas S1, Rand MD, Lake RJ (1999) Notch signaling: cell fate control and signal integration in development. Science 284: 770-776.
-
Gao J, Chen C, Hong L, Wang J, Du Y, et al. (2007) Expression of Jagged1 and its association with hepatitis B virus X protein in hepatocellular carcinoma. Biochem Biophys Res Commun 356: 341-347.
-
Giovannini C, Lacchini M, Gramantieri L, Chieco P, Bolondi L (2006) Notch3 intracellular domain accumulates in HepG2 cell line. Anticancer Res 26: 2123-2127.
-
Gonsalves FC, Weisblat DA (2007) MAPK regulation of maternal and zygotic Notch transcript stability in early development. Proc Natl Acad Sci U S A 104: 531-536.
-
Stockhausen MT, Sjölund J, Axelson H (2005) Regulation of the Notch target gene Hes-1 by TGFalpha induced Ras/MAPK signaling in human neuroblastoma cells. Exp Cell Res 310: 218-228.
-
Chappell WH, Green TD, Spengeman JD, McCubrey JA, Akula SM, et al. (2005) Increased protein expression of the PTEN tumor suppressor in the presence of constitutively active Notch-1. Cell Cycle 4: 1389-1395.
-
Reya T, Clevers H (2005) Wnt signalling in stem cells and cancer. Nature 434: 843-850.
-
Gordon MD, Nusse R (2006) Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem 281: 22429-22433.
-
Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW (1997) Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem 272: 24735-24738.
-
Merle P, de la Monte S, Kim M, Herrmann M, Tanaka S, et al. (2004) Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology 127: 1110-1122.
-
Inagawa S, Itabashi M, Adachi S, Kawamoto T, Hori M, et al. (2002) Expression and prognostic roles of beta-catenin in hepatocellular carcinoma: correlation with tumor progression and postoperative survival. Clin Cancer Res 8: 450-456.
-
Joo M, Lee HK, Kang YK (2003) Expression of beta-catenin in hepatocellular carcinoma in relation to tumor cell proliferation and cyclin D1 expression. J Korean Med Sci 18: 211-217.
-
Laurent-Puig P, Legoix P, Bluteau O, Belghiti J, Franco D, et al. (2001) Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology 120: 1763-1773.
-
Taniguchi K, Roberts LR, Aderca IN, Dong X, Qian C, et al. (2002) Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene 21: 4863-4871.
-
Yamashita T, Budhu A, Forgues M, Wang XW (2007) Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res 67: 10831-10839.
-
Yang W, Yan HX, Chen L, Liu Q, He YQ, et al. (2008) Wnt/beta-catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells. Cancer Res 68: 4287-4295.
-
Mishra L, Derynck R, Mishra B (2005) Transforming growth factor-beta signaling in stem cells and cancer. Science 310: 68-71.
-
Weinstein M, Yang X, Deng C (2000) Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev 11: 49-58.
-
Mishra L, Cai T, Yu P, Monga SP, Mishra B (1999) Elf3 encodes a novel 200-kD beta-spectrin: role in liver development. Oncogene 18: 353-364.
-
Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, et al. (2003) Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science 299: 574-577.
-
Breuhahn K, Longerich T, Schirmacher P (2006) Dysregulation of growth factor signaling in human hepatocellular carcinoma. Oncogene 25: 3787-3800.
-
Tsai JF, Jeng JE, Chuang LY, Yang ML, Ho MS, et al. (1997) Elevated urinary transforming growth factor-beta1 level as a tumour marker and predictor of poor survival in cirrhotic hepatocellular carcinoma. Br J Cancer 76: 244-250.
-
Bedossa P, Peltier E, Terris B, Franco D, Poynard T (1995) Transforming growth factor-beta 1 (TGF-beta 1) and TGF-beta 1 receptors in normal, cirrhotic, and neoplastic human livers. Hepatology 21: 760-766.
-
Yuan F, Zhou W, Zou C, Zhang Z, Hu H, et al. (2010) Expression of Oct4 in HCC and modulation to wnt/? -catenin and TGF-? signal pathways. Mol Cell Biochem 343: 155-162.
-
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663-676.
-
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861-872.
-
Chow EK, Fan LL, Chen X, Bishop JM (2012) Oncogene-specific formation of chemoresistant murine hepatic cancer stem cells. Hepatology 56: 1331-1341.
-
Machida K, Tsukamoto H, Mkrtchyan H, Duan L, Dynnyk A, et al. (2009) Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci U S A 106: 1548-1553.
-
Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, et al. (2009) Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460: 1132-1135.
-
Yin X, Li YW, Zhang BH, Ren ZG, Qiu SJ, et al. (2012) Coexpression of stemness factors Oct4 and Nanog predict liver resection. Ann Surg Oncol 19: 2877-2887.
-
Rountree CB, Mishra L, Willenbring H (2012) Stem cells in liver diseases and cancer: recent advances on the path to new therapies. Hepatology 55: 298-306.
-
Ning N, Pan Q, Zheng F, Teitz-Tennenbaum S, Egenti M, et al. (2012) Cancer stem cell vaccination confers significant antitumor immunity. Cancer Res.72, 1853-1864.
-
Rountree CB, Mishra L, Willenbring H (2012) Stem cells in liver diseases and cancer: recent advances on the path to new therapies. Hepatology 55: 298-306.
-
Ning N, Pan Q, Zheng F, Teitz-Tennenbaum S, Egenti M, et al. (2012) Cancer stem cell vaccination confers significant antitumor immunity. Cancer Res.72, 1853-1864.