International Journal of Clinical Biostatistics and Biometrics
Bayesian Survival Analysis of Genetic Variants in PTPRN2 Gene for Age at Onset of Cancer
Ke-Sheng Wang1*, Yue Pan2, Weize Wang3 and Chun Xu4
1Department of Biostatistics and Epidemiology, East Tennessee State University, USA
2Department of Public Health Sciences, University of Miami, USA
3Department of Biostatistics, Florida International University, USA
4Department of Pediatrics, Texas Tech University Health Sciences Center, USA
*Corresponding author:
Kesheng Wang, Department of Biostatistics and Epidemiology, College of Public Health, East Tennessee State University, PO Box 70259, Lamb Hall, Johnson City, TN 37614-1700, USA, Tel:1-423-439-4481, Fax: 1- 423-439- 4606, E-mail: wangk@etsu.edu
Int J Clin Biostat Biom, IJCBB-1-004, (Volume 1, Issue 1), Research Article; ISSN: 2469-5831
Received: August 12, 2015 | Accepted: September 07, 2015 | Published: September 09, 2015
Citation: Wang KS, Pan Y, Wang W, Xu C (2015) Bayesian Survival Analysis of Genetic Variants in PTPRN2 Gene for Age at Onset of Cancer. Int J Clin Biostat Biom 1:004. 10.23937/2469-5831/1510004
Copyright: © 2015 Wang KS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: The protein tyrosine phosphatase, receptor type, N polypeptide 2 (PTPRN2) gene may play a role in cancer; however, no study has focused on the associations of genetic variants within the PTPRN2 gene with age at onset (AAO) of cancer.
Methods: This study examined 220 single nucleotide polymorphisms (SNPs) within the PTPRN2 gene in the Marshfield sample with 716 cancer cases (any diagnosed cancer, excluding minor skin cancer) and 2,848 non-cancer controls. Multiple logistic regression model and linear regression model in PLINK software were used to examine the association of each SNP with the risk of cancer and AAO, respectively. For survival analysis of AAO, both classic Cox regression and Bayesian survival analysis using the Cox proportional hazards model in SAS v. 9.4 were applied to detect the association of each SNP with AAO. The hazards ratios (HRs) with 95% confidence intervals (CIs) were estimated.
Results: Single marker analysis identified 10 SNPs associated with the risk of cancer and 9 SNPs associated with AAO (p < 0.05). SNP rs7783909 revealed the strongest association with cancer (p = 6.52x10-3); while the best signal for AAO was rs4909140 (p = 6.18x10-4), which was also associated with risk of cancer (p = 0.0157). Classic Cox regression model showed that 11 SNPs were associated with AAO (top SNP rs4909140 with HR = 1.38, 95% CI = 1.11-1.71, p = 3.3x10-3). Bayesian Cox regression model showed similar results to those using the classic Cox regression (top SNP rs4909140 with HR = 1.39, 95% CI = 1.1-1.69).
Conclusions: This study provides evidence of several genetic variants within the PTPRN2 gene influencing the risk of cancer and AAO, and will serve as a resource for replication in other populations.
Keywords
Cancer, Age at onset, PTPRN2, Single nucleotide polymorphism, Bayesian analysis, Survival analysis
Introduction
Cancer continues to remain a significant public health issue globally. It is the leading cause of death in both developed and emerging economies [1]. In 2012, there were 14.1 million new cancer cases, 8.2 million cancer mortalities, and 32.6 million people living with cancer globally [2]. Cancers are caused by a complex interplay between genetic predisposition and environment. Family and twin studies have shown the estimated effects of heritability of the colorectal cancer (35%) [3], breast cancer (25-30%) [3-5], prostate cancer (42-58%) [3,6], and lung cancer (25-26%) [3,7].
The protein tyrosine phosphatase, receptor type, N polypeptide 2 (PTPRN2) gene (also known as IAR, ICAAR, PTPRP, IA-2beta, R-PTP-N2) is located at 7q36 [8,9]. The PTPRN2 is expressed primarily in human brain and pancreas and in mouse brain, pancreas, and insulinoma cell lines [10,11]. One methylation study found that PTPRN2 showed highly significant hypermethylation in squamous cell lung cancer tissue [12]. Another gene expression profile study suggested that PTPRN2 was associated with metastatic prostate cancer [13]. A recent study reported that PTPRN2 was expressed predominantly in endocrine and neuronal cells, where it might function in exocytosis and suggested as a novel candidate biomarker and therapeutic target in breast cancer [14].
However, no study has focused on the associations of genetic variants of PTPRN2 gene with age at onset (AAO) of cancer. Furthermore, Bayesian methods have become increasingly popular in many areas of scientific research including genetic association studies, which may have some advantages in flexibility, and incorporating information from previous studies and dealing with sparse-data [15-17]. In this study, we explored the associations of 220 SNPs in the PTPRN2 gene with AAO of cancer using a Bayesian survival analysis in a Caucasian sample.
Subjects and Methods
The Marshfield sample
The Marshfield sample is from the publicly available data in A Genome-Wide Association Study on Cataract and HDL in the Personalized Medicine Research Project Cohort - Study Accession: phs000170.v1.p1 (dbGaP). The details about these subjects were described elsewhere [18,19]. Cancer cases were defined as any diagnosed cancer excluding minor skin cancer; while AAO cancer was defined by date of the earliest cancer diagnosis in the registry. Social factors used in this study were age, gender, alcohol use in the past month (yes or no), and smoking status (never smoking, current smoking and past smoking). Obesity was determined as a body mass index (BMI) ≥ 30. Genotyping data using the ILLUMINA Human660W-Quad_v1_A are available for 3564 Caucasian individuals (716 cancer cases and 2848 controls). Within the PTPRN2 gene, 220 SNPs were available and therefore included in the analysis.
Statistical methods
Linear and logistic regression models in PLINK software: The categorical variables were presented as frequencies and percentages. The continuous variables were reported as the means ± standard deviation. Quality-control and association analyses were implemented using PLINK V1.07 [20]. First, Hardy-Weinberg equilibrium (HWE) was tested for all the SNPs using the controls; then, minor allele frequency (MAF) was determined for each SNP. Multiple logistic regression analysis of each SNP with risk of cancer as a binary trait, adjusted for sex, age*age, alcohol use, smoking status, and obesity, was performed using PLINK; while the asymptotic p-values were observed and the odds ratio (OR) and 95% confident interval (CI) were estimated. Furthermore, AAO values were firstly log transformed, then multiple linear regression analysis of each SNP with log transformed AAO of cancer, adjust for sex, alcohol use, smoking status, and obesity was performed; while the asymptotic p-values were observed and the regression coefficient (β) and 95% CI were estimated. To control for type I errors arising from multiple hypothesis testing, a false discovery rate (FDR) was defined in Benjamini and Hochberg [21] as the expected proportion of false discoveries. In addition, empirical p-values were generated by 100,000 permutation tests using Max (T) permutation procedure. In this procedure, pointwise estimate of an individual SNP's significance (empirical pointwise p-values) was calculated.
Cox proportional hazards models in PROC PHREG: The proportional hazards model or Cox regression model, is widely used in the analysis of time-to-event data to explain the effect of explanatory variables on hazard rates. The PHREG procedure fits the Cox model by maximizing the partial likelihood function; this eliminates the unknown baseline hazard and accounts for censored survival times. In the Bayesian approach, the partial likelihood function is used as the likelihood function in the posterior distribution [22]. In the non-Bayesian analysis, the Akaike information criterion (AIC) was used as a measure of goodness of model fit that balances model fit against model simplicity [23,24]. Bayesian Cox regression can be requested by using the BAYES statement in the PHREG procedure. A Markov chain Monte Carlo (MCMC) method by Gibbs sampling was used to simulate samples from the posterior distribution. In a Bayesian analysis, a Gibbs chain of samples from the posterior distribution was generated for the model parameters. Summary statistics (mean, standard deviation, quartiles, the highest posterior density (HPD) and credible intervals, correlation matrix) and convergence diagnostics (Geweke; the effective sample size; and Monte Carlo standard errors) were computed for each parameter, as well as the correlation matrix and the covariance matrix of the posterior sample. Trace plots, posterior density plots, and autocorrelation function plots were created for each parameter [17].
For the present study of the AAO, the normal prior was chosen for the coefficients. In Bayesian analysis, a deviance information criterion (DIC) is available for model comparison instead of AIC. DIC is a hierarchical modeling generalization of the AIC; while DIC is intended as a generalization of AIC [25]. The following program showed one SNP rs4909140, sex, alcohol use, smoking status, and obesity with the AAO of cancer. The rs4909140 has 3 genotypes - G_G, G_T and T_T, respectively; while the T_T genotype was considered as the reference.
proc phreg data= aao;
class sex(ref ="1") obesity (ref ="1") alcohol(ref ="1") smoking(ref ="1") rs4909140 (ref = "T_T") ;
model canceraao*statuscan(0) = sex obesity alcohol smoking rs4909140/risklimits;
bayes seed =1000 nbi =10000 nmc =100000 thin =10 seed =1000 cprior=normal(var =1e6) outpost = out plots = density;
hazardratio rs4909140;
run;
Descriptive statistics and Cox regression analyses were conducted with SAS v.9.4 (SAS Institute, Cary, NC, USA).
Results
Genotype quality control and descriptive statistics
We removed 1 SNP with HWE p < 10-4. All other 219 SNPs were in HWE with MAF > 1% in the controls. The demographic characteristics of the subjects in the study are presented in Table 1. There were slightly more females than males in both cases and controls. The age ranged from 46 to 90 years and AAO of cancer ranged from 23 to 90 years.
![]()
Table 1: Descriptive characteristics of cases and controls
View Table 1
Multiple linear and logistic regression analyses using PLINK
Using a single marker analysis, we identified 10 SNPs associated with the risk of cancer and 9 SNPs associated with AAO (p < 0.05) in the Marshfield sample (Table 2). SNP rs7783909 revealed the strongest association with cancer (p = 6.52x10-3); while the best signal for AAO was rs4909140 (p = 6.18x10-4), which was also associated with risk of cancer (p = 0.0157). For the 10 SNPs associated with risk of cancer, the FDR was 90%; while the FDR for the two AAO mostly associated SNPs (rs4409140 and rs1670340) were 21% and 39%, respectively. Furthermore, we conducted a permutation test in PLINK and found that all the cancer and or AAO associated SNPs had empirical point wise p-values p < 0.05 using a permutation test (Table 2).
![]()
Table 2: SNPs within the PTPRN2 gene associated with risk and age at onset of cancer using PLINK (p < 0.05)
View Table 2
Classic and Bayesian Cox regression analyses using PROC PHREG
Classic Cox regression model showed that 11 SNPs were associated with AAO (top SNP rs4909140 with HR=1.38, 95%CI = 1.11-1.71, p = 3.3x10-3). The HRs based on the Bayesian survival analyses revealed similar results to those using the non-Bayesian analyses results (Table 3). The DIC for the 11 SNPs in the Bayesian analyses were similar to those of AIC using classic Cox model.
![]()
Table 3: SNPs within the PTPRN2 gene associated with AAO of cancer using PROC PHREG (p < 0.05)
View Table 3
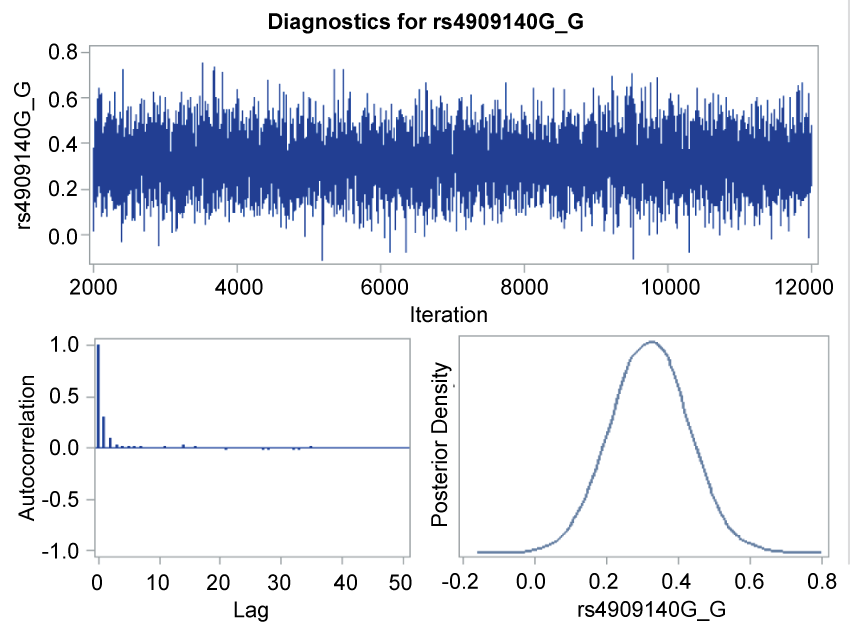
.
Figure 1: Trace plot, autocorrelation function plot, and posterior density plot for rs4909140.
View Figure 1
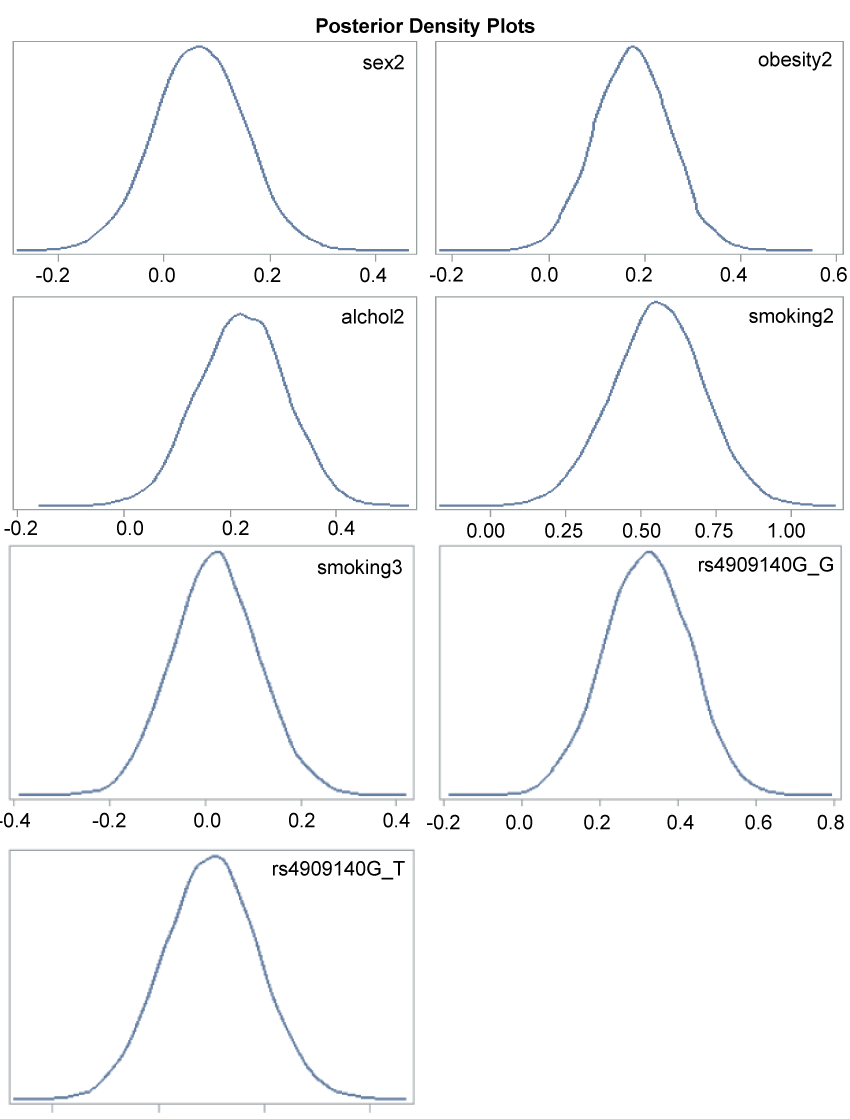
The trace plot, posterior density plot, and autocorrelation function plot based on Bayesian analysis (Figure 1) indicated that the Markov chain had stabilized with good mixing for rs4909140. The posterior density plot, which estimate the posterior marginal distributions for the 7 regression coefficients showed a smooth, unimodal shape for the posterior marginal distribution (Figure 2). Table 4 shows the posterior summary of rs4909140 with HR = 1.39, 95% CI = 1.1-1.69.
![]()
Table 4: Posterior summary and hazard ratio for rs4909140
View Table 4
Discussion
In this study, we identified 10 SNPs associated with the risk of cancer and 9 SNPs with AAO using the PLINK software and 11 SNPs revealed associations with AAO using Cox survival model in SAS. Bayesian Cox regression model revealed similar findings to those using the classic Cox regression. To our knowledge, this is the first candidate gene study to provide evidence of several genetic variants within the PTPRN2 gene associated with the risk of cancer and AAO.
Previous studies have showed that PTPRN2 is an autoantigen for type 1 diabetes which is an insulin-dependent diabetes mellitus and autoimmune disease; while PTPRN2 is reactive with type 1 diabetes patient sera and is likely to be an islet cell antigen useful in the preclinical screening of individuals for the risk of type 1 diabetes [10,26-28]. Animal model studies revealed that this gene may be functioned in the regulation of insulin secretion [11,29-32]. Recently, another study suggested that PTPRN2 (IA-2beta) is one of the genes potentially relevant to insulin and neurotransmitter release [33]. Furthermore, several studies have reported that the PTPRN2 gene may be involved in squamous cell lung cancer tissue [12], metastatic prostate cancer [13] and breast cancer [14]. However, the mechanism is not clear. It has been hypothesized that recurrent or clonal somatic mutation underlies the initiation of autoimmune disease such as type 1 diabetes [34]; while many cancers elicit antibodies that are also found in autoimmune diseases [35]. Therefore, PTPRN2 may be one of the mechanisms linking autoimmune diseases to cancers. In addition, insulin, insulinlike growth factor 1, and insulinlike growth factor 2 signaling through the insulin receptor and the insulinlike growth factor 1 receptor could induce tumorigenesis, accounting to some extent for the link between diabetes, metabolic syndrome and cancers [36,37].
However, no association study of genetic polymorphisms within the PTPRN2 gene with the risk of cancer and AAO has been conducted. The present study provides the first evidence of several genetic variants within the PTPRN2 gene is associated with the risk and AAO of cancer using multiple logistic and linear regression models. We identified the main effects and permutation p-values for single SNPs. Furthermore, we conducted Bayesian survival analysis of genetic variants with AAO. Bayesian methods may have some advantages in flexibility and incorporating information from previous studies. For example, Bayesian method may provide an alternative approach to assessing associations that alleviates the limitations of p-values at the cost of some additional modelling. It has recently made great inroads into many areas of science, including the assessment of associations between genetic variants and disease or related phenotypes [15]. We also realized some limitations in this study. First, the definition of cancer status in the Marshfield sample was broad (including any diagnosed cancer omitting minor skin cancer). It would be more informative to investigate the association of PTPRN2 gene with particular types of cancer. Furthermore, our current findings might be subject to type I error and findings need to be replicated in additional samples.
Conclusion
This study provides evidence of several genetic variants within the PTPRN2 gene influencing the risk and AAO of cancer. Future functional study of this gene may help to better characterize the genetic architecture of cancers.
Role of the funding sources
No founding source is given for the present paper.
Disclosure
All authors have reported no financial interests or potential conflicts of interest.
Acknowledgement
Funding support for the Personalized Medicine Research Project (PMRP) was provided through a cooperative agreement (U01HG004608) with the National Human Genome Research Institute (NHGRI), with additional funding from the National Institute for General Medical Sciences (NIGMS). The samples used for PMRP analyses were obtained with funding from Marshfield Clinic, Health Resources Service Administration Office of Rural Health Policy grant number D1A RH00025, and Wisconsin Department of Commerce Technology Development Fund contract number TDF FYO10718. Funding support for genotyping, which was performed at Johns Hopkins University, was provided by the NIH (U01HG004438). Assistance with phenotype harmonization and genotype cleaning was provided by the eMERGE Administrative Coordinating Center (U01HG004603) and the National Center for Biotechnology Information (NCBI). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000170.v1.p1. This study was approved by the Internal Review Board (IRB), East Tennessee State University.
References
-
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, et al. (2015) Global cancer statistics, 2012. CA Cancer J Clin 65: 87-108.
-
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, et al. (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136: E359-386.
-
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, et al. (2000) Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343: 78-85.
-
Czene K, Lichtenstein P, Hemminki K (2002) Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer 99: 260-266.
-
Locatelli I, Lichtenstein P, Yashin AI (2004) The heritability of breast cancer: a Bayesian correlated frailty model applied to Swedish twins data. Twin Res 7: 182-191.
-
Hjelmborg JB, Scheike T, Holst K, Skytthe A, Penney KL, et al. (2014) The heritability of prostate cancer in the Nordic Twin Study of Cancer. Cancer Epidemiol Biomarkers Prev 23: 2303-2310.
-
Jin Y, Zhou X, He X (2001) [The general measurement of genetic factors on lung cancer in Xuanwei, China]. Zhongguo Fei Ai Za Zhi 4: 354-356.
-
Smith PD, Barker KT, Wang J, Lu YJ, Shipley J, et al. (1996) ICAAR, a novel member of a new family of transmembrane, tyrosine phosphatase-like proteins. Biochem Biophys Res Commun 229: 402-411.
-
Morahan G, Huang D, Yu WP, Cui L, DeAizpurua H, et al. (1998) Localization of the genes encoding the type I diabetes autoantigens, protein-tyrosine phosphatases IA2 and IAR. Mamm Genome 9: 593-594.
-
Cui L, Yu WP, DeAizpurua HJ, Schmidli RS, Pallen CJ (1996) Cloning and characterization of islet cell antigen-related protein-tyrosine phosphatase (PTP), a novel receptor-like PTP and autoantigen in insulin-dependent diabetes. J Biol Chem 271: 24817-24823.
-
Doi A, Shono T, Nishi M, Furuta H, Sasaki H, et al. (2006) IA-2beta, but not IA-2, is induced by ghrelin and inhibits glucose-stimulated insulin secretion. Proc Natl Acad Sci U S A 103: 885-890.
-
Anglim PP, Galler JS, Koss MN, Hagen JA, Turla S, et al. (2008) Identification of a panel of sensitive and specific DNA methylation markers for squamous cell lung cancer. Mol Cancer 7: 62.
-
Chen CL, Mahalingam D, Osmulski P, Jadhav RR, Wang CM, et al. (2013) Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate 73: 813-826.
-
Sorokin AV, Nair BC, Wei Y, Aziz KE, Evdokimova V, et al. (2015) Aberrant Expression of proPTPRN2 in Cancer Cells Confers Resistance to Apoptosis. Cancer Res 75: 1846-1858.
-
Stephens M, Balding DJ (2009) Bayesian statistical methods for genetic association studies. Nat Rev Genet 10: 681-690.
-
Sullivan SG, Greenland S (2013) Bayesian regression in SAS software. Int J Epidemiol 42: 308-17.
-
Stokes M, Chen F, Gunes F (2014) An introduction to Bayesian analysis with SAS/STAT® software. Proceedings of the SAS Global Forum 2014 Conference, SAS Institute Inc, Cary, NC.
-
McCarty CA, Peissig P, Caldwell MD, Wilke RA (2008) The Marshfield Clinic Personalized Medicine Research Project: 2008 scientific update and lessons learned in the first 6 years. Personalized Medicine 5: 529-542.
-
McCarty CA, Wilke RA, Giampietro PF, Wesbrook SD, Caldwell MD (2005) Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Medicine 2: 49-79.
-
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559-575.
-
Benjamini Y, and Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist Soc 57: 289-300.
-
Sinha D, Ibrahim JG, and Chen M (2003) Bayesian Justification of Cox's Partial Likelihood. Biometrics 90: 629-641.
-
Akaike H (1979) A Bayesian Extension of the Minimum AIC Procedure of Autoregressive Model Fitting. Biometrika 66: 237-242.
-
Akaike H (1981) Likelihood of a Model and Information Criteria. Journal of Econometrics 16:3-14.
-
Spiegelhalter DJ, Best NG, Carlin BP, Van der Linde A (2002) Bayesian Measures of Model Complexity and Fit (with Discussion). J. R. Statist. Soc. B 64: 583-616.
-
Kawasaki E, Hutton JC, Eisenbarth GS (1996) Molecular cloning and characterization of the human transmembrane protein tyrosine phosphatase homologue, phogrin, an autoantigen of type 1 diabetes. Biochem Biophys Res Commun 227: 440-447.
-
Lu J, Li Q, Xie H, Chen ZJ, Borovitskaya AE, et al. (1996). Identification of a second transmembrane protein tyrosine phosphatase, IA-2beta, as an autoantigen in insulin-dependent diabetes mellitus: precursor of the 37-kDa tryptic fragment. Proc Natl Acad Sci U S A 93: 2307-2311.
-
Li Q, Borovitskaya AE, DeSilva MG, Wasserfall C, Maclaren NK, et al. (1997) Autoantigens in insulin-dependent diabetes mellitus: molecular cloning and characterization of human IA-2 beta. Proc Assoc Am Physicians 109: 429-439.
-
Cui L, Yu WP, Pallen CJ (1998) Insulin secretagogues activate the secretory granule receptor-like protein-tyrosine phosphatase IAR. J Biol Chem 273: 34784-34791.
-
Löbner K, Steinbrenner H, Roberts GA, Ling Z, Huang GC, et al. (2002) Different regulated expression of the tyrosine phosphatase-like proteins IA-2 and phogrin by glucose and insulin in pancreatic islets: relationship to development of insulin secretory responses in early life. Diabetes 51: 2982-2988.
-
Kubosaki A, Gross S, Miura J, Saeki K, Zhu M, et al. (2004) Targeted disruption of the IA-2β gene causes glucose intolerance and impairs insulin secretion but does not prevent the development of diabetes in NOD mice. Diabetes 53: 1684-1691.
-
Xu H, Abuhatzira L, Carmona GN, Vadrevu S, Satin LS, et al. (2015) The Ia-2β intronic miRNA, miR-153, is a negative regulator of insulin and dopamine secretion through its effect on the Cacna1c gene in mice. Diabetologia .
-
Mandemakers W, Abuhatzira L, Xu H, Caromile LA, Hébert SS, et al. (2013) Co-regulation of intragenic microRNA miR-153 and its host gene Ia-2β: identification of miR-153 target genes with functions related to IA-2β in pancreas and brain. Diabetologia 56: 1547-1556.
-
Ross KA (2014) Coherent somatic mutation in autoimmune disease. PLoS One 9: e101093.
-
Bei R, Masuelli L, Palumbo C, Modesti M, Modesti A (2009) A common repertoire of autoantibodies is shared by cancer and autoimmune disease patients: Inflammation in their induction and impact on tumor growth. Cancer Lett 281: 8-23.
-
Gallagher EJ, Fierz Y, Ferguson RD, LeRoith D (2010) The pathway from diabetes and obesity to cancer, on the route to targeted therapy. Endocr Pract 16: 864-873.
-
Gallagher EJ, LeRoith D (2013) Epidemiology and molecular mechanisms tying obesity, diabetes, and the metabolic syndrome with cancer. Diabetes Care 36 Suppl 2: S233-239.