Journal of Genetics and Genome Research
Relationship of Mitochondrial DNA Haplogroups with Complex Diseases
Carlos G Urzua-Traslavina1 , Maria G Moreno-Trevino1, Denisse A Martinez-Trevino1, Hugo A Barrera-Saldana2 and Rafael BR Leon-Cachon1*
1Universidad de Monterrey, Division Ciencias de la Salud, Departamento de Ciencias Basicas, San Pedro Garza García, Nuevo Leon, Mexico
2Universidad Autonoma de Nuevo Leon, Facultad de Medicina, Monterrey, Nuevo Leon, Mexico
*Corresponding author:
Rafael Baltazar Reyes Leon-Cachon, Av. Ignacio Morones Prieto 4500 Pte., Jesus M. Garza, San Pedro Garza Garcia, Nuevo Leon, 66238, Mexico, Tel: +528182151448; Fax: +5283337747; E-mail: rafael.reyesleon@udem.edu/yagami_rleon@hotmail.com
J Genet Genome Res, JGGR-1-011, (Volume 1, Issue 2), Review Article; ISSN: 2378-3648
Received: November 28, 2014 | Accepted: December 13, 2014 | Published: December 16, 2014
Citation: Urzua-Traslavina CG, Moreno-Trevino MG, Martínez-Trevino DA, BarreraSaldana HA, Leon-Cachon RBR (2014) Relationship of Mitochondrial DNA Haplogroups with Complex Diseases. J Genet Genome Res 1:011. 10.23937/2378-3648/1410011
Copyright: © 2014 Urzua-Traslavina CG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Mitochondria are responsible for energy production in unicellular and multicellular eukaryotes. Apart from their major role in metabolism, mitochondria are involved in many other cellular processes. A new paradigm is needed to understand aging, aging-related illnesses, and complex diseases. Because of the long co-evolution of mitochondria with the cells, subtle variations in the function of these organelles could influence many organ systems. Understanding the extent of this influence will shed light on the pathophysiology of some of these diseases. This review will examine the history of mitochondrial DNA research, and explain the role mitochondrial DNA plays in relation to complex diseases and their outcomes, like metabolic diseases, cancer and obesity. Finally, we will explore some of the new paradigms used in this research.
Keywords
Mitochondrial DNA, Haplogroups, Obesity, Complex diseases, Association, Therapy response
Introduction
The mitochondrion is a special organelle because it is the main producer of cellular energy, has its own DNA (mtDNA), transcription and replication machinery [1]. The mitochondrial genome has substantial similarity to prokaryotic genomes. Therefore, it is believed that the mitochondrion is the remnant of an ancient symbiosis between a prokaryote and a eukaryote. First, the prokaryotic symbiont was incorporated into the eukaryotic cell as an organelle. Next, the majority of the prokaryotic genes (1500 approximately) moved into the nucleus of human cells with only 37 genes remain inside the mitochondrion [2]. Therefore, mitochondria and cell have become interdependent that nuclear genes involved in mitochondrial function must evolve more quickly to keep up with the mutation rate of mtDNA. critical for optimal mitochondrial function and consequently affects human health [3].
More recently, the role of mitochondria in other cellular processes has been. For example, in the regulation of the cytoplasmic calcium level [4], apoptosis [5], and signal transduction [6]. mitochondria influence the human phenotype to a higher extent than previously assumed [7], since subtle variations in cell metabolism affect different organ systems in complex ways [8]. In addition, mitochondria play an important role in the aging process [9]. In this article we explore to what extent the analysis of mtDNA can help us understand certain complex diseases.
Early mtDNA Research
MtDNA possesses certain attributes that makes it ideal for specific applications. Early mtDNA research focused on retracing the steps our ancestors took when migrating out of Africa and into the other continents [10]. This research for example benefitted from the negligible recombination and faster mutation rate of mtDNA, which resulted in an increased resolution in the analysis of the ancestral lineages. Moreover, mtDNA has become a powerful tool in the forensic analysis of partially degraded samples because of its higher copy number in comparison to nuclear DNA [11].
This research also led to some discoveries in the field of medicine, when mutations on the coding mtDNA were found to be responsible for non-Mendelian hereditary diseases, which are known nowadays as mitochondrial diseases [12]. Efforts then focused on understanding how mutations in mtDNA arise, how they are selected in the cell population and how they are passed on to the offspring. answers to these questions the phenomenon of mitochondrial heteroplasmy, which is partially responsible for the clinical variability observed in mitochondrial diseases [13]. Heteroplasmy occurs when mitochondrial genomes mutate during embryogenesis, which causes the formation of a variety of lineages that coexist in the same individual [14]. The intracellular accumulation of mutant mtDNA on this stage occurs by a poorly understood selection mechanism. Then, the asymmetrical distribution of mutated mitochondria during mitosis affects mitochondrial function in the daughter cells, and the tissues they will form, sometimes involving the germinal cell line and eventually fixating on the population through genetic drift and/or selection [15]. After gestation, the selection process varies according to the organ system involved. Although many organ systems are quite compliant [16], accumulation of mutated mtDNA scarcely occurs in certain cell types. For example, mice studies suggest that oocytes are selected based on their mitochondrial competence [17]. Conversely, bone marrow cells are subject to a selective pressure that avoids mutations which compromise the efficiency of energy production [18. Depending on the time of the mutation and the rate of cellular turnover, even a mild genetic variation on a single mitochondrion may significantly impact the function of many tissues [19].
The Relationship between MtDNA and Complex Diseases
As MtDNA mutates more quickly than nuclear DNA [3], natural selection may shape the effect mitochondria have on metabolism and even on the functionality of organ systems on a shorter amount of time. For example, the mitochondrial efficiency hypothesis proposes that in warm climate adapted populations, the evolution of mtDNA favored the conversion of energy to adenosine triphosphate (ATP) rather than to heat. Such mitochondrial adaptations may be disadvantageous in modern western lifestyles characterized by physical inactivity and high calorie diets [20]. Thus, the mitochondrial efficiency hypothesis could help explain the increased susceptibility to diabetes observed in south Asian populations [21]. Likewise, in cold climate adapted populations, mtDNA favors heat production at the expense of efficient energy conversion, increasing the likelihood that the next mutation could severely compromise the cell's energy production. This could explain why some populations like northern Europeans are more susceptible to mitochondrial diseases [22] (Figure 1).
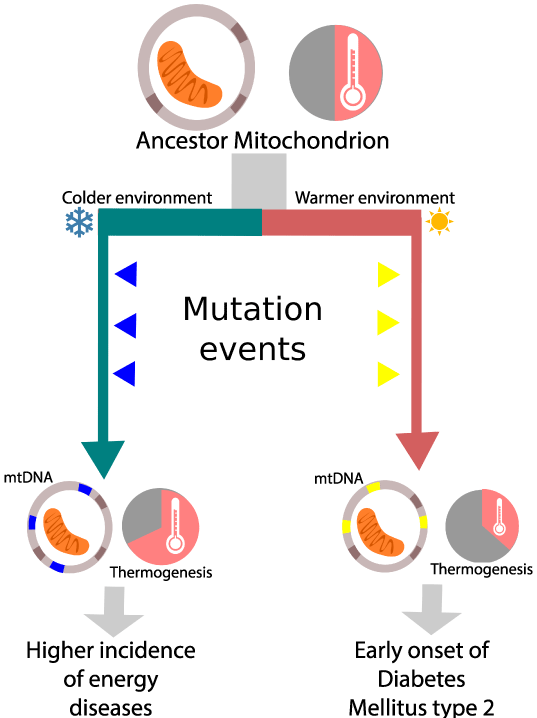 Figure 1: The mitochondrial efficiency hypothesis proposes that mitochondrial lineages evolving in different environmental conditions will be shaped differently by natural selection, resulting in variations in their mitochondrial function. Those variations could explain the increased incidence of certain types of diseases. Using the mutation pattern they acquired during evolution, associations with such diseases may be made.
View Figure 1
Figure 1: The mitochondrial efficiency hypothesis proposes that mitochondrial lineages evolving in different environmental conditions will be shaped differently by natural selection, resulting in variations in their mitochondrial function. Those variations could explain the increased incidence of certain types of diseases. Using the mutation pattern they acquired during evolution, associations with such diseases may be made.
View Figure 1
This information led to the hypothesis that although mtDNA can affect more than one organ system subtler mitochondrial variations may be more difficult to detect phenotypically. Therefore, with the advances in sequencing technologies new research focused on searching associations between mtDNA and the so called "complex" diseases, known to involve multiple organ systems, and of elusive etiology [19]. Because mitochondria play a key role in metabolism, diseases like diabetes, metabolic syndrome and obesity were initially investigated. Later on, it was found that slight defects in the mitochondrial repair system could also help explain the etiology of neurodegenerative diseases like Alzheimer [23] and Parkinson's disease [24]. Furthermore, mtDNA is explored in relation to cancer because of the role mitochondria play in the regulation of apoptosis [25].
Currently, mtDNA sequences can be classified according to the haplogroups they belong to. Members of a haplogroup have one or more distinct SNP's in common, which they all inherited from a common ancestor. The haplogroups are named according to a letter of the alphabet, but their lexicographical order guards no relation to their genetic relationships. The most recent common ancestor of all mitochondria belongs to haplogroup L. This haplogroup gave rise to seven distinct haplogroups (L0-L6), which are currently located in Africa. The group of humans that migrated out of Africa belonged to haplogroup L3, consequently all non-African haplogroups come from this group alone. Haplogroup L3 gave rise to the M and N subclades. In particular, European haplogroups (I, J, K, T, U, V, W, H, U, and X) emerged from the N subclade. On the other hand, Asian haplogroups developed from both the N subclade (A, B, F, and Y) and the M subclade (C, D, G, Q, and Z). Finally, during the migration to the Americas, haplogroups C and D (from the M subclade) and haplogroups A, B and X, (N subclade) became the predominant genotypes in this continent [26]. Thus, each haplogroup represents a major branching point in the evolutionary tree of the mitochondrial genome.
Haplogroups help distinguish populations that have evolved independently during recent human history [10]. Because the resolution of the haplogroups is limited by the mitochondrial rate of mutation, the SNPs located in the mitochondrial control region are especially useful because this region's mutation rate is even faster than the rest of the mtDNA [27]. Consequently, if some diseases are affected by mitochondrial function, they should behave differently depending on the haplogroup of the patient.
In particular, haplogroup N9a was found to be significantly related to resistance to metabolic syndrome in Japanese women [28]. In southern Italy, haplogroup T was found to be a risk factor for morbid obesity [body mass index (BMI) >45kg/m2] [29]. In a Taiwanese population, haplogroup B4 was associated with diabetes [30]. And in a Caucasian population, haplogroup J increased the risk of age related macular degeneration [31] (Table 1).
![]()
Table 1: Associations between mitochondrial haplogroups and mtDNA mutations with diseases.
View Table 1
Actually, high throughput DNA analysis tools, like microarrays and next generation sequencing are used to verify or reject the aforementioned associations. Additionally, new insights into the levels of heteroplasmy (which turned out to be higher than previously thought) [14], and the screening of larger sample sizes improved research quality and statistical power.
For example, full mtDNA sequences could be mined from an exome data set composed of 1000 individuals with type 2 diabetes and 1000 controls from a Danish population. Among the 2025 polymorphisms that were found to be associated to diabetes type 2, 393 had not been reported before [32]. A group in Germany reported no association between BMI and coding mtDNA using an array of 119 SNPs. But based on their results they recommended further analysis of control region variants [33]. Interestingly, another team found that some polymorphisms in the coding regions were associated with BMI, using a larger sample size [34].
The potential of these polymorphisms to be of any use in the treatment and understanding of these diseases remains to be seen. Currently, the main obstacle to mtDNA research is low sample sizes , because of high sequencing costs. Hopefully, better analysis will be possible in the near future as the amount of data on genome increases thanks to next generation sequencing technologies.
The Relationship between mtDNA and Disease Outcomes
One of the first clinical uses of mtDNA was the identification of mutations responsible for mitochondrial diseases. Apart from helping in diagnosis, knowledge of the mutations did not improve treatment options significantly. Treatment for these diseases still relies heavily on symptomatic control, changes in diet and exercises regimens [35]. Nevertheless, data obtained so far about the frequency and location of the affecting mutations have provided some insights into these diseases. For example, LOHN progression results in visual loss due to neuropathy more frequently in haplogroup J in carriers with the 11778 G to A or the 14484 T to C mutation [36]. Haplogroups have also been associated with outcomes in other non-mitochondrial diseases. Haplogroups H and R were found to be an independent predictor of survival in patients with severe sepsis [37,38]. Similarly, haplogroup T was found to be protective of lipoatrophy after highly active antiretroviral therapy [39]. Even though most of these findings require further validation, the potential role of mtDNA in the stratification of patients cannot be ignored.
One of the most controversial topics is the link between mtDNA and cancer. In 1920, the Nobel laureate Otto Heinrich Warburg hypothesized that tumorigenesis was the result of insufficient cellular respiration caused by mitochondrial damage. He postulated this after observing the anaerobic metabolism cancer cells possess even in the presence of oxygen (Warburg effect). Modern research has now revealed how little we knew about the role of mitochondrial function in cancer. It is now known that the anaerobic metabolism in cancer cells is not a result of mtDNA damage, but a well regulated metabolic reprogramming [40]. Additionally, mitochondrial function was recently shown to vary between epithelial and stromal breast cancer cells [41]. Moreover, a non-intuitive result revealed colon cancer cells to have a slower mutation rate than normal tissue [42].
Clearly, because of the complexity of cancer cells, the analysis of their mtDNA is bound to be plagued with difficulties. For example, rapid tumor growth could be facilitated by an mtDNA mutation. This mutation could become ubiquitous in the tumor cells because of clonal selection. Nevertheless, using a mathematical model, one study reported that non-functional mutations could become established similarly during rapid tumor cell growth entirely by chance [43]. Despite the difficulties just mentioned, another group found mitochondrial mutations to be responsible for the modulation of metastatic potential in breast cancer cells [44], this was only accomplished by observing a decrease in metastatic potential after replacing the mitochondria inside the cancer cells with mitochondria from healthy cells.
Even though the analysis of mtDNA inside cancer cells is still rudimentary, cancer incidence and progression could still be associated to mitochondrial haplogroups in a similar fashion as with other diseases. Efforts into this area have found an increased predisposition for breast and esophageal cancer in Haplogroup N [45]. Also, haplogroup U was found to increase the risk of prostate and renal cancer in a North American population [46]. Another research team associated the North East Asian haplogroup CZD (Comprised of M8a, C, Z, D4 and D5) with good disease-free survival in oral squamous cell carcinoma patients [47].
The Way we See Diseases Frames the Way we do Research
Justifiably, many of the previous association research efforts have focused on one specific disease, and as such established inclusion, exclusion, and other criteria to enable association for the disease of interest. However, this may not be the best approach, because the classification criteria possibly encompass many different diseases that share the same clinical outcome. For example, different forms of Parkinson have distinct etiology but similar clinical features [48]. Also the pathophysiology of a disease may cause other diseases, for example, obesity leads to diabetes and hypertension [49]. The aforementioned has to be taken into account when designing these studies.
Following this line of thinking, associations have been found between mtDNA polymorphisms and groups of diseases rather of only a single disease [50]. This idea came from the multiorgan effects characteristic of mitochondrial diseases, and the insights provided by the research of human mitochondrial heteroplasmy conducted so far [14].
Another approach is to sub-classify diseases according to disease progression. For example, bacterial infections are classified according to their antibiotic resistance [51]. Similarly, associations between mtDNA and AIDS progression were found in a European population. Haplogroups which had less efficient mitochondrial function characterized by more thermogenesis (U5 and J), were associated with accelerated disease progression, and more efficient groups characterized by lower heat production (H3, H4, H5 and H6) were associated with disease protection [52]. suggests that differences in mitochondrial function may affect the evolution of some diseases.
Mitochondrial Function and Obesity
The World Health Organization defines obesity as the abnormal or excessive fat accumulation that may impair health. Clinically a BMI of over 30 defines obesity [53].
Nowadays, obesity has become an alarming health problem in the new century [54]. Current treatment for obese patients include recommendations for lifestyle changes, and diets which may be complemented with pharmacological treatment, performing bariatric surgery in difficult cases [55]. Weight loss often involves a serious commitment, sometimes with suboptimal results, and surely tests the level of trust and communication between patient and physician.
Moderate weight loss benefits the prognosis of patients with metabolic syndrome, insulin resistance, hypertension [56] and diabetes [57]. This is the main reason behind the understanding of obesity as a disease with future complications. Even so, a high BMI could be nothing more than a non-pathognomonic clinical indicator of disease, like fever in other diseases [58]. If it were found that obese patients responded in different ways to a standard weight loss strategy, the classification into treatment response groups would be justified and it would be possible to investigate the role the pathophysiology of obesity plays on these differences, giving additional options for prevention and treatment of these patients.
To our knowledge, the effect of diets on human mitochondrial function has not been studied so far. However, it has been reported that the liver mitochondria of mice maintained with calorie restriction acquired more cristae [59]. Another work found a change in mitochondrial number in orexigenic neurons from mice during the transition from fasted to fed state [60].
Following this line of, we are performing a pilot study to find associations between mtDNA and the response to calorie restriction diet in an overweight human population. We expect to discover polymorphisms that can predict the success of such diet which would be a useful biomarker in the stratification of obese patients undergoing treatment.
Conclusion
Mitochondria are unusual organelles with unorthodox genomics and a long history of co-evolution with eukaryotes. Subtle changes in their function could help explain the variability of complex diseases across populations and also the relationship with some neurodegenerative diseases that appear later in life. Mitochondria can be readily distinguished based on the analysis of mtDNA and such differentiation can be associated with functioning of the organism. Nevertheless, mtDNA research is still limited by lack of data and more time is needed to become useful as a biomarker.
Acknowledgement
The authors thank Irene Meester for reviewing this manuscript.
Conflicts of interest
We confirm that there are no known conflicts of interest associated with this publication and the financial support for this work has not influenced its outcome.
References
-
Bonawitz ND, Clayton DA, Shadel GS (2006) Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell 24: 813-825.
-
Lang BF, Gray MW, Burger G (1999) Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet 33: 351-397.
-
Gershoni M, Levin L, Ovadia O, Toiw Y, Shani N, et al. (2014) Disrupting mitochondrial-nuclear coevolution affects OXPHOS complex I integrity and impacts human health. Genome Biol Evol 6: 2665-2680.
-
Demaurex N, Poburko D, Frieden M (2009) Regulation of plasma membrane calcium fluxes by mitochondria. Biochim Biophys Acta 1787: 1383-1394.
-
Wang C, Youle RJ (2009) The role of mitochondria in apoptosis*. Annu Rev Genet 43: 95-118.
-
Kluge MA, Fetterman JL, Vita JA (2013) Mitochondria and endothelial function. Circ Res 112: 1171-1188.
-
Larsen FJ, Schiffer TA, Sahlin K, Ekblom B, Weitzberg E, et al. (2011) Mitochondrial oxygen affinity predicts basal metabolic rate in humans. FASEB J 25: 2843-2852.
-
Wallace DC (2013) A mitochondrial bioenergetic etiology of disease. J Clin Invest 123: 1405-1412.
-
Terman A, Kurz T, Navratil M, Arriaga EA, Brunk UT (2010) Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxid Redox Signal 12: 503-535.
-
Soares P, Alshamali F, Pereira JB, Fernandes V, Silva NM, et al. (2012) The Expansion of mtDNA Haplogroup L3 within and out of Africa. Mol Biol Evol 29: 915-927.
-
Zapico SC, Ubelaker DH (2013) mtDNA Mutations and Their Role in Aging, Diseases and Forensic Sciences. Aging Dis 4: 364-380.
-
Dimauro S (2011) A history of mitochondrial diseases. J Inherit Metab Dis 34: 261-276.
-
Mroczek-Tońska K, Kisiel B, Piechota J, Bartnik E (2003) Leber hereditary optic neuropathy--a disease with a known molecular basis but a mysterious mechanism of pathology. J Appl Genet 44: 529-538.
-
Ramos A, Santos C, Mateiu L, Gonzalez Mdel M, Alvarez L, et al. (2013) Frequency and pattern of heteroplasmy in the complete human mitochondrial genome. PLoS One 8: e74636.
-
Wallace DC, Chalkia D (2013) Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol 5: 021220.
-
He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, et al. (2010) Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 464: 610-614.
-
Dalton CM, Carroll J (2013) Biased inheritance of mitochondria during asymmetric cell division in the mouse oocyte. J Cell Sci 126: 2955-2964.
-
't Hart LM, Jansen JJ, Lemkes HH, de Knijff P, Maassen JA (1996) Heteroplasmy levels of a mitochondrial gene mutation associated with diabetes mellitus decrease in leucocyte DNA upon aging. Hum Mutat 7: 193-197.
-
Wallace DC (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39: 359-407.
-
Bhopal RS, Rafnsson SB (2009) Could mitochondrial efficiency explain the susceptibility to adiposity, metabolic syndrome, diabetes and cardiovascular diseases in South Asian populations? Int J Epidemiol 38: 1072-1081.
-
Bakker LE, Sleddering MA, Schoones JW, Meinders AE, Jazet IM (2013) Pathogenesis of type 2 diabetes in South Asians. Eur J Endocrinol 169: R99-99R114.
-
Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC (2004) Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303: 223-226.
-
Swerdlow RH (2012) Mitochondria and cell bioenergetics: increasingly recognized components and a possible etiologic cause of Alzheimer's disease. Antioxid Redox Signal 16: 1434-1455.
-
Exner N, Lutz AK, Haass C, Winklhofer KF (2012) Mitochondrial dysfunction in Parkinson's disease: molecular mechanisms and pathophysiological consequences. EMBO J 31: 3038-3062.
-
Birch-Machin MA (2006) The role of mitochondria in ageing and carcinogenesis. Clin Exp Dermatol 31: 548-552.
-
Maca-Meyer N, González AM, Larruga JM, Flores C, Cabrera VM (2001) Major genomic mitochondrial lineages delineate early human expansions. BMC Genet 2: 13.
-
Galtier N, Jobson RW, Nabholz B, Glémin S, Blier PU (2009) Mitochondrial whims: metabolic rate, longevity and the rate of molecular evolution. Biol Lett 5: 413-416.
-
Tanaka M, Fuku N, Nishigaki Y, Matsuo H, Segawa T, et al. (2007) Women with mitochondrial haplogroup N9a are protected against metabolic syndrome. Diabetes 56: 518-521.
-
Nardelli C, Labruna G, Liguori R, Mazzaccara C, Ferrigno M, et al. (2013) Haplogroup T is an obesity risk factor: mitochondrial DNA haplotyping in a morbid obese population from southern Italy. Biomed Res Int 2013: 631082.
-
Liou CW, Chen JB, Tiao MM, Weng SW, Huang TL, et al. (2012) Mitochondrial DNA coding and control region variants as genetic risk factors for type 2 diabetes. Diabetes 61: 2642-2651.
-
Mueller EE, Schaier E, Brunner SM, Eder W, Mayr JA, et al. (2012) Mitochondrial haplogroups and control region polymorphisms in age-related macular degeneration: a case-control study. PLoS One 7: e30874.
-
Li S, Besenbacher S, Li Y, Kristiansen K, Grarup N, et al. (2014) Variation and association to diabetes in 2000 full mtDNA sequences mined from an exome study in a Danish population. Eur J Hum Genet 22: 1040-1045.
-
Knoll N, Jarick I, Volckmar AL, Klingenspor M, Illig T, et al. (2014) Mitochondrial DNA variants in obesity. PLoS One 9: e94882.
-
Flaquer A, Baumbach C, Kriebel J, Meitinger T, Peters A, et al. (2014) Mitochondrial genetic variants identified to be associated with BMI in adults. PLoS One 9: e105116.
-
Parikh S, Saneto R, Falk MJ, Anselm I, Cohen BH, et al. (2009) A modern approach to the treatment of mitochondrial disease. Curr Treat Options Neurol 11: 414-430.
-
Man PY, Howel N, Mackey DA, Nørby S, Rosenberg T, et al. (2004) Mitochondrial DNA haplogroup distribution within Leber hereditary optic neuropathy pedigrees. J Med Genet 41: e41.
-
Baudouin SV, Saunders D, Tiangyou W, Elson JL, Poynter J, et al. (2005) Mitochondrial DNA and survival after sepsis: a prospective study. Lancet 366: 2118-2121.
-
Yang Y, Shou Z, Zhang P, He Q, Xiao H, et al. (2008) Mitochondrial DNA haplogroup R predicts survival advantage in severe sepsis in the Han population. Genet Med 10: 187-192.
-
Hendrickson SL, Kingsley LA, Ruiz-Pesini E, Poole JC, Jacobson LP, et al. (2009) Mitochondrial DNA haplogroups influence lipoatrophy after highly active antiretroviral therapy. J Acquir Immune Defic Syndr 51: 111-116.
-
Ward PS, Thompson CB (2012) Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21: 297-308.
-
Sanchez-Alvarez R, Martinez-Outschoorn UE, Lamb R, Hulit J, Howell A, et al. (2013) Mitochondrial dysfunction in breast cancer cells prevents tumor growth: understanding chemoprevention with metformin. Cell Cycle 12: 172-182.
-
Ericson NG, Kulawiec M, Vermulst M, Sheahan K, O'Sullivan J, et al. (2012) Decreased mitochondrial DNA mutagenesis in human colorectal cancer. PLoS Genet 8: e1002689.
-
Coller HA, Khrapko K, Bodyak ND, Nekhaeva E, Herrero-Jimenez P, et al. (2001) High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat Genet 28: 147-150.
-
Imanishi H, Hattori K, Wada R, Ishikawa K, Fukuda S, et al. (2011) Mitochondrial DNA mutations regulate metastasis of human breast cancer cells. PLoS One 6: e23401.
-
Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN (2007) Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett 249: 249-255.
-
Booker LM, Habermacher GM, Jessie BC, Sun QC, Baumann AK, et al. (2006) North American white mitochondrial haplogroups in prostate and renal cancer. J Urol 175: 468-472.
-
Lai CH, Huang SF, Chen IH, Liao CT, Wang HM, et al. (2012) The mitochondrial DNA Northeast Asia CZD haplogroup is associated with good disease-free survival among male oral squamous cell carcinoma patients. PLoS One 7: e49684.
-
Schapira AH (2009) Etiology and pathogenesis of Parkinson disease. Neurol Clin 27: 583-603, v.
-
Bray GA (2004) Medical consequences of obesity. J Clin Endocrinol Metab 89: 2583-2589.
-
Hudson G, Gomez-Duran A, Wilson IJ, Chinnery PF (2014) Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet 10: e1004369.
-
Davies J, Davies D (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74: 417-433.
-
Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, et al. (2008) Mitochondrial DNA haplogroups influence AIDS progression. AIDS 22: 2429-2439.
-
WHO. Fact sheet N°311: Obesity and overweight.
-
Wyatt SB, Winters KP, Dubbert PM (2006) Overweight and obesity: prevalence, consequences, and causes of a growing public health problem. Am J Med Sci 331: 166-174.
-
Hainer V, Toplak H, Mitrakou A (2008) Treatment modalities of obesity: what fits whom? Diabetes Care 31 Suppl 2: S269-277.
-
Pasanisi F, Contaldo F, de Simone G, Mancini M (2001) Benefits of sustained moderate weight loss in obesity. Nutr Metab Cardiovasc Dis 11: 401-406.
-
Fujioka K (2010) Benefits of moderate weight loss in patients with type 2 diabetes. Diabetes Obes Metab 12: 186-194.
-
Harrington M, Gibson S, Cottrell RC (2009) A review and meta-analysis of the effect of weight loss on all-cause mortality risk. Nutr Res Rev 22: 93-108.
-
Khraiwesh H, Lopez-Domínguez JA, Lopez-Lluch G, Navas P, de Cabo R, et al. (2013) Alterations of ultrastructural and fission/fusion markers in hepatocyte mitochondria from mice following calorie restriction with different dietary fats. J Gerontol A Biol Sci Med Sci 68: 1023-1034.
-
Dietrich MO, Liu ZW, Horvath TL (2013) Mitochondrial dynamics controlled by mitofusins regulate Agrp neuronal activity and diet-induced obesity. Cell 155: 188-199.
-
Fernández-Caggiano M, Barallobre-Barreiro J, Rego-Pérez I, Crespo-Leiro MG, Paniagua MJ, et al. (2012) Mitochondrial haplogroups H and J: risk and protective factors for ischemic cardiomyopathy. PLoS One 7: e44128.
-
Wolf C, Gramer E, Müller-Myhsok B, Pasutto F, Wissinger B, et al. (2010) Mitochondrial haplogroup U is associated with a reduced risk to develop exfoliation glaucoma in the German population. BMC Genet 11: 8.
-
Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, et al. (1988) Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 242: 1427-1430.
-
Solano A, Roig M, Vives-Bauza C, Hernandez-Peña J, Garcia-Arumi E, et al. (2003) Bilateral striatal necrosis associated with a novel mutation in the mitochondrial ND6 gene. Ann Neurol 54: 527-530.
-
Silvestri G, Ciafaloni E, Santorelli FM, Shanske S, Servidei S, et al. (1993) Clinical features associated with the A->G transition at nucleotide 8344 of mtDNA ("MERRF mutation"). Neurology 43: 1200-1206.
-
Goto Y, Nonaka I, Horai S (1990) A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348: 651-653.