Journal of Genetics and Genome Research
Experimental Detection of Mitochondrial DNA Insertions in Nuclear Genome of Chicken Embryos Developed from X-Ray Irradiated Eggs
Serazhutdin A. Abdullaev* and Azhub I. Gaziev
Institute of Theoretical and Experimental Biophysics, Russian Academy of Sciences, Pushchino, Russian Federation
*Corresponding author:
Serazhutdin A. Abdullaev, Institute of Theoretical and Experimental Biophysics of the Russian Academy of Sciences, Pushchino, Moscow Region, 142290, Russian Federation, Tel: +007-4967-739364; E-mail: saabdullaev@gmail.com
J Genet Genome Res, JGGR-1-008, (Volume 1, Issue 2), Research Article; ISSN: 2378-3648
Received: November 22, 2014 | Accepted: December 06, 2014 | Published: December 08, 2014
Citation: Abdullaev SA, Gaziev AI (2014) Experimental Detection of Mitochondrial DNA Insertions in Nuclear Genome of Chicken Embryos Developed from X-Ray Irradiated Eggs. J Genet Genome Res 1:008. 10.23937/2378-3648/1410008
Copyright: © 2014 Abdullaev SA. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The transfer of mitochondrial DNA (mtDNA) into the nuclear genome is a dynamic process, resulting in the formation of nuclear mitochondrial (numt) pseudogenes or numt-insertions. Experimental determination of de novo numt-insertions is limited by the extensive homology of mtDNA in the nuclear DNA (nDNA) of eukaryotes. Since chicken nDNA contains only 13 numt-pseudogenes, we tried to follow experimentally the induction of numt-insertions de novo in the nDNA of chicken (Gallus gallus) embryos developed from eggs subjected to X-ray irradiation. NDNA of chicken embryo liver were twice purified from free mtDNA by gel-electrophoresis and monitored by PCR. PCR were run to determine the numt-insertions in the nDNA of surviving embryos, using 11 primer pairs flanking regions of mtDNA size of 300-400 bp. However, the PCR of control group nDNA, by using the given primers, revealed no homology with mtDNA. PCR of nDNA of embryos from irradiated eggs testified the origination of amplified mtDNA regions in two among eight embryos. Two and three loci of mtDNA were reproducibly identified in purified nDNA from two individual embryos. The sequencing of PCR amplicons synthesized from these nDNA matrices showed that they were identical to mtDNA. Thus the results indicate that ionizing radiation can induce integration of mtDNA fragments into the nuclear genome, perhaps in the process of repair of double strand breaks in nDNA via a non-homologous end-joining mechanism. However, it can be assumed that the insertion of large fragments of mtDNA in nuclear genome, as in this experiment, is a rare event.
Keywords
Induction numt-insertions, Ionizing radiation, Chicken embryos
Introduction
The transfer of mitochondrial genetic material into the nucleus and its integration in the nuclear genome is commonly believed to be a continuous and dynamic process. Fragments of mitochondrial DNA (mtDNA) in the nuclear genome are found as non-coding sequences, known as nuclear mitochondrial (numt) pseudogenes or numt-insertions [1-6]. The localization of numt-pseudogenes in the nuclear genome is currently studied in many higher organisms from yeasts to humans. The numt-pseudogenes are distributed over different chromosomes, persist in the genome as "fossil molecular elements" and form a "library" of mtDNA fragments that have migrated into the nuclear genome, thus providing a highly significant information on the history of genome evolution [2,4-6]. Numt-insertions can be not only considered as neutral polymorphic sites but are often associated with carcinogenesis, aging and genetic diseases in humans [7-12].
The incorporation of mtDNA fragments into the nuclear genome requires their escape from mitochondria. This may occur due to mtDNA damage, destruction of mitochondria, or in the process of division and mitophagy of these organelles [2,13-15]. Blanchard and Schmidt [16] hypothesized that mtDNA fragments can integrate into the nuclear genome during the reunion of broken chromosomal ends. This assumption was supported by a number of studies, giving grounds for a possibility of repair of double-strand breaks (DSB) of nuclear DNA (nDNA) accompanied by "capturing" of mtDNA fragments through non-homologous end-joining (NHEJ), and also with participation of microhomology regions on terminal sequences [5,17-21]. These points to the dependence of de novo numt-pseudogene formation frequency on the rate of occurrence of DSBs in nDNA, the activity of their repair by the NHEJ mechanism, as well as on the amount of mtDNA fragments migrating into the nucleus from organelles. We have previously proposed that such events are likely to occur following the influence of ionizing radiation on the organism, which induces the damage of mtDNA and nDNA with formation of DSB [21]. However, the existent literature on this subject lacks experimental confirmation of numt-pseudogene formation de novo in the genomes of organisms subjected to ionizing radiation. Experimental identification of numt-insertions de novo is difficult because of numerous regions of mtDNA homology constitutively present in eukaryotic nuclear genomes. We made an attempt of experimental detection of de novo origination of extensive numt-insertions in the nDNA of chicken (Gallus gallus) embryo liver obtained from X-ray irradiated eggs. The realization of analogous experiments using mouse models or human cells has been unsuccessful because of the high density of numt-pseudogenes in their nuclear genomes. Our choice of chicken as an object for study was dictated by that the chicken genome originally contains only 13 numt-insertions (0.0001% of the genome size) [5,21], while mouse and human genomes have 190 and 871 numt insertions, respectively [5]. We show that numt-pseudogenes arise de novo in the nuclear genome of chicken embryos obtained from X-ray irradiated eggs.
Materials and Methods
Chicken eggs and their irradiation
Fertilized eggs from chicken (Gallus gallus domesticus) of the White Leghorn chicken breed were obtained from a poultry farm (Tula, Russia). Two days after laying, the eggs were placed in plastic containers (5 eggs in each) and subjected to X-ray radiation. Irradiation was carried out using an X-ray unit TU-12 ("Medrent", Russia) at 280 kV, 18 mA, with a dose rate of 1 Gy/min. The absorbed dose was 5 Gy. Immediately after irradiation, the eggs were put for incubation (two groups of 10 irradiated and control eggs) at standard temperature and humidity. After 19 days of incubation, the eggs were opened. The irradiated group had 8 live embryos, while all the10 embryos survived in the control group.
Isolation and purification of nuclear DNA
Liver samples were taken from embryos, cleaned from membranes and homogenized in buffer A (10 mmol/l Tris-HCl, pH 8.0, 0.5 mol/l sucrose, 25 mmol/l KCl, 10 mmol/l MgCl2, 2.5% NP-40) at the ratio 1:3 at 2-3°Cin a Dounce homogenizer. The obtained homogenate was 3-fold diluted with buffer A, filtered through a capron mesh and centrifuged at 1000xg for 15 min. The nuclei-containing sediment was washed with buffer A by resuspending and centrifuging. After purification, the nuclei were resuspended in lysis buffer (10 mmol/l Tris-HCl, pH 8.0, 1mmol/l EDTA, 0.5% SDS, 20mkg/ml RNase) and incubated at 37°C for 45 min. DNA was subsequently isolated using a standard phenol-chloroform technique. The obtained samples were purified by gel-electrophoresis in 0.7% agarose to remove free mtDNA. Following the first electrophoresis, agarose strips containing high molecular nDNA were cut out from the gel and purified by electrophoresis for the second time. Areas of agarose gel containing high molecular DNA were visualized with ethidium bromide using a UV transilluminator. DNA was extracted from agarose gel and its subsequent purification was done with "DNA Cleanup" (Bio-Rad Laboratories, Hercules, USA) according to recommendations of the manufacturer. The amount of DNA was measured by reaction with Pico Green, according to the manufacturer's protocol (Molecular Probes, Eugene, OR, USA) and fluorescence was registrated using Tecan Infinite 200 (Austria).
PCR of long DNA fragments (long-extension PCR)
PCR of long fragments (long-extension PCR) was carried out with DNA samples (the first fraction was isolated from nuclei and electrophoretically purified fractions) to evaluate the impurities of free mtDNA. An mtDNA region of 15495 bp (covering more than 92% of the mitochondrial genome) was amplified and flanked by primers H1255 (5'-CATCTTGGCATCTTCAGTGCC-3') and L16750 (5'-AGGACTACGGCTTGAAAAGC-3') used in [23]. PCR of a part of the gene β-actin (3193 bp) with primers For 5'-ACAATGGCTCCGGTATGTGC AA-3', Rev 5'-CTGTAAAGCCTTCATTCACATCTAT-3' [24] was used as a positive control for PCR with nDNA. PCR of long mtDNA fragments and the β-actin-coding gene was realized in the same tube. The PCR reaction mixture (a total volume of 25 μl) contained: 75 mM/l Tris-HCI, pH 8.8, 20 mM/l (NH4)2SO4, 2.5 mM/l MgCI2, 200 μM/l of each dNTP, 250 nM/l of each primer, 0.01% Tween-20. The reaction mixture was supplemented with 5 ng of DNA and 1.5 U of a mixture of Taq and Pfu DNA polymerases (Thermo Scientific, USA). The enzyme mixture was added for the "hot start" after the initial denaturation of the DNA template at 94°C for 4 min. PCR was run for 40 cycles of: 1 min denaturation at 94°C, 30 s annealing at 64°C and 5 min elongation at 72°C, followed by the final 10 min incubation at 72°C. PCR products were run in 0.8% agarose gel. Synthesis of all primers, including those listed in Table 1, was done by "Syntol" (Moscow, Russia). In all cases, the PCR was run on a programmed thermal cycler "Tertsik" ("DNA Technology", Moscow, Russia). PCR products were visualized on a UV transilluminator following gel electrophoresis with ethidium bromide.
PCR of mtDNA sequences using nDNA template
PCR was carried out using purified nDNA samples isolated from liver from both groups of embryos. A total of 11 pairs of primers were used, which corresponded to mtDNA and the flanking regions of 300-400 bp (Table). These primers were taken from [25,26]. The reaction mixture (25 μl), including primers and purified nDNA (5 ng), contained the same components, which were used for PCR of long fragments of mtDNA. PCR was run for 40 cycles: 30s denaturation at 94°C, 30s annealing at 62°Cand 30s elongation at 72°C, followed by the final 4 min incubation at 72°C. The products of PCR amplification were analyzed electrophoretically in 1% agarose gel.
Sequencing PCR products
Products of PCR amplification of mtDNA regions were extracted from agarose gel and purified using a "DNA Cleanup" kit ("Bio-Rad", USA), as mentioned above. The nucleotide sequences of PCR-derived amplicons were done according to the method of Sanger (Sanger F). A Reagent kit ABI-PRISM(γ)BigDye™ Terminator V.3.1 (GE Healthcare, USA) was used, with a subsequent analysis of reaction products using an automated DNA sequencer ABI-PRISM 3730 (Applied Biosystems, USA). The sequences of interest were analyzed in both directions. The obtained reads of direct and reverse strands were compared pairwise to exclude the possible mistakes of sequencing.
Results
Experiments for identification of numt-pseudogenes originated de novo in the nuclear genomes of animal tissues require a thorough purification of analyzed nDNA samples free from impurities of mtDNA. For this reason, all samples of liver nDNA from each embryo were twice purified by gel-electrophoresis and tested for absence of free mtDNA impurities by the methods short and long extension PCR (Figure 1). Figure 1° C represents three electrophoregrams of nuclear DNA isolated from chicken embryo liver. The first lane shows DNA obtained by a standard phenol-chloroform technique from lysed nuclei, without further electrophoretic purification, lanes 2 and 3 - samples resulting from electrophoretic separation of highest molecular fractions cut out from the gel for subsequent electrophoresis. It is clearly seen that the double electrophoresis of nDNA samples results in the removal of low-molecular impurities (Figure 1° C, lane 3). The purified fractions of nDNA from each embryo were tested by PCR for the presence of impurities of free mtDNA, using primers flanking the extensive regions of mtDNA (15495 bp) and nDNA (β-actin 3193 bp) (Figure 1B). On the gel, it is seen that DNA samples isolated from nuclei (lane 1) contain impurities of mtDNA. However, it is obvious that using nDNA, twice purified electrophoretically, as a template for PCR gives no amplicons corresponding to mtDNA products (Figure 1B, lane 3). This means that double electrophoresis was successful for purification of nDNA samples from long fragments of free mtDNA. PCR analysis, involving nDNA as templates and 11 pairs of primers for mtDNA fragments, testified the presence of free mtDNA impurities in most nDNA samples of the first fraction (Figure 1C). Identical PCR tests (with 11 primer pairs) of electrophoretically purified samples of liver nDNA from chicken embryos taken out of unirradiated eggs gave negative results (Figure 2°C). The obtained data prove that double electrophoresis allowed efficient purification of nDNA samples from free mtDNA fragments. Inserts of mtDNA were revealed in purified nDNA samples of embryos from 10 unirradiated eggs and of embryos from 8 irradiated eggs. PCR were run with each nDNA sample and with all 11 primer pairs for 3-4 times. Testing of 10 nDNA templates from control embryos with the abovementioned 11 primer pairs revealed no mtDNA sequences in them (Figure 2°C exemplifies the results of such PCR analysis for one nDNA sample). At the same time, PCR with 11 primer pairs using nDNA templates isolated from irradiated egg embryos revealed amplified regions in two nDNA samples only. Stable detection was observed for PCR products from two and three loci of mtDNA in the nDNA samples of embryos No 6 and No 7, respectively.
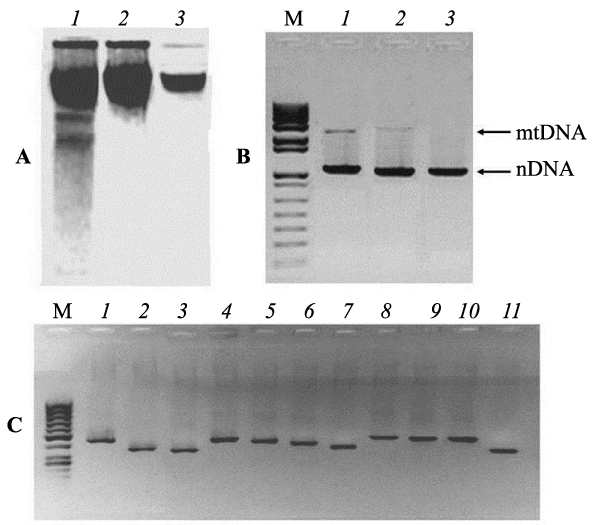 Figure 1: Purification of nDNA samples and testing the presence of impurities of free mtDNA by PCR. A-purification of nDNA by double gel-electrophoresis: 1, initial nDNA from nuclei of embryo liver; 2, after the first electrophoresis; 2, after the second electrophoresis. B- Gel-electrophoresis of PCR products of extensive mtDNA and nDNA regions (β-actin gene). Lane 1- initial nDNA from embryo liver nuclei, lanes 2 and 3- the same DNA samples after the first and the second rounds of purification by gel-electrophoresis. C: Gelelectrophoresis of PCR products obtained from nDNA isolated from liver nuclei (without electrophoretic purification) with 11 primer pairs listed in Table 1. M: DNA length markers.
View Figure 1
Figure 1: Purification of nDNA samples and testing the presence of impurities of free mtDNA by PCR. A-purification of nDNA by double gel-electrophoresis: 1, initial nDNA from nuclei of embryo liver; 2, after the first electrophoresis; 2, after the second electrophoresis. B- Gel-electrophoresis of PCR products of extensive mtDNA and nDNA regions (β-actin gene). Lane 1- initial nDNA from embryo liver nuclei, lanes 2 and 3- the same DNA samples after the first and the second rounds of purification by gel-electrophoresis. C: Gelelectrophoresis of PCR products obtained from nDNA isolated from liver nuclei (without electrophoretic purification) with 11 primer pairs listed in Table 1. M: DNA length markers.
View Figure 1
Figure 2 shows electrophoregrams of PCR products synthesized from purified nDNA of embryos taken out of irradiated eggs (B - embryo No 6, C- embryo No 7). Numbers on the top part of electrophoregrams correspond to the primer pairs listed in Table 1. Lane C(control) corresponds to the PCR product obtained from electrophoretically unpurified nDNA of an unirradiated egg using primer pair 5 (Table 1). Figure 2 shows that the amplification of two mtDNA regions initiated by 10 and 11 pairs of primers occurs on the matrix of nDNA of embryo number 6. The amplification of three mtDNA regions initiated by 3, 8 and 11pairs of primers occurs on the matrix of the nDNA of embryo number 7. No synthesis was observed on the other six embryo nDNA samples from irradiated eggs (data not shown). These results indicate that the integration of mtDNA fragments into nDNA of liver took place in only two of the eight survived embryos from irradiated eggs. PCR- amplicons synthesized on nDNA templates of embryos from irradiated eggs (No 6 and No 7) were excised from agarose gel, purified and sequenced as described earlier. The results of analysis are presented in Fig 3, where only the sequences of the L-strand of mtDNA are shown. Analyses show that the nucleotide sequences of amplicons are identical to mtDNA and to the sites of genes indicated (Figure 3). The sites of 12S rRNA and 16S rRNA genes are localized in the nDNA from embryo No 6, and the nDNA from embryo No 7 contains the sequences of COI, ND5 and 12S rRNA genes of mtDNA (Figure 3). Nucleotide sequence analysis also showed that the observed numt-insertions in nDNA contain rare single nucleotide polymorphisms that are not present in the homologous sites of free mtDNA. These polymorphisms are due to two or three substitutions of individual bases or deletions in the sequences of numt-inserts.
![]()
Table 1: Primers used in PCR for determination of mtDNA insertions in the nuclear genome of chicken embryo liver.
View Table 1
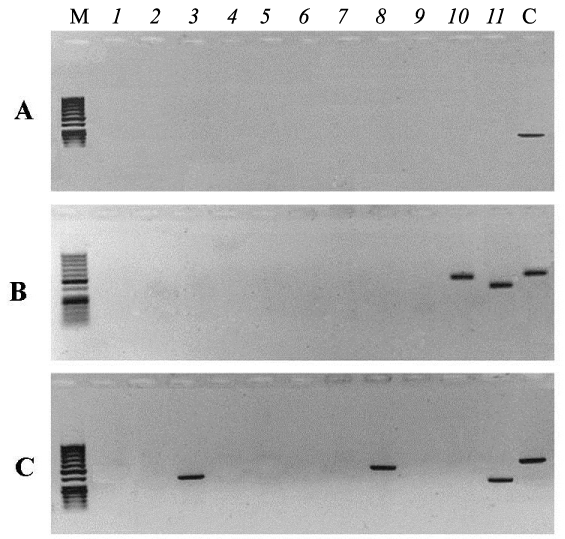 Figure 2: Gel-electrophoresis of PCR products from the templates of nDNA of embryos from unirradiated eggs (A) and embryos No 6 and No 7 from irradiated eggs (B, C), twice purified by electrophoresis. Lanes (1-11) - PCR with primer pairs listed in Table 1. C - control, PCR product from unpurified nDNA and primer pair 5 (Table 1). M: DNA length marker.
View Figure 2
Figure 2: Gel-electrophoresis of PCR products from the templates of nDNA of embryos from unirradiated eggs (A) and embryos No 6 and No 7 from irradiated eggs (B, C), twice purified by electrophoresis. Lanes (1-11) - PCR with primer pairs listed in Table 1. C - control, PCR product from unpurified nDNA and primer pair 5 (Table 1). M: DNA length marker.
View Figure 2
Thus, the results of this experiment confirm the assumption of the possibility of induction of numt-insertions into the nuclear genome of cells exposed to ionizing radiation. However, it can be assumed that the insertion of large fragments of mtDNA (300-400 bp) in nuclear genome, as in this experiment, is a rare event.
 Figure 3: Nucleotide sequences (L-strands) of PCR amplicons synthesized from purified nDNA templates of embryo No 6 (12SrRNA, 16SrRNA) and No 7 (12SrRNA, COI, ND5) from irradiated eggs. Sequences which are underlined and marked with arrows correspond to direct and reverse primers (Table 1).
View Figure 3
Figure 3: Nucleotide sequences (L-strands) of PCR amplicons synthesized from purified nDNA templates of embryo No 6 (12SrRNA, 16SrRNA) and No 7 (12SrRNA, COI, ND5) from irradiated eggs. Sequences which are underlined and marked with arrows correspond to direct and reverse primers (Table 1).
View Figure 3
Discussion
Detection of large mtDNA sequences in the nuclear genome of chicken embryos developed from X-ray irradiated eggs indicates that in cells damaged by radiation, conditions are created for the formation of de novo numt-pseudogenes in nDNA. However, the insertions of mtDNA into liver nDNA were registered not in all embryos developed from irradiated eggs. Probably it depended on the conditions of the experiment, - first of all, on a limited number of chosen mtDNA sites and their large size (300-400 bp) for PCR amplification. Perhaps the amplification of small sizes mtDNA we could reveal more numt-inserts in the nDNA. In any case, the insertion of mtDNA fragments into nDNA sequences is presumably an infrequent event. It is also possible that the integration of mtDNA into the gene-coding nDNA sequences that are determinative of the development and survival of the embryo is lethal.
Analysis of literature data suggests that the frequency of "capturing" mtDNA by the nuclear genome depends on the rate of DSB occurring in nDNA, the activity of NHEJ repair systems, as well as on the amount of mtDNA fragments migrating into the nucleus [2,5,17-21]. Such a situation, beneficial for integration of mtDNA fragments into the nuclear genome, is developed following the exposure of the organism to some damaging agents, predominantly ionizing radiation [13,21]. MtDNA is currently considered to be a more sensitive target for different damaging factors compared to nDNA. For example, it has been previously demonstrated, that mtDNA gets more damage from ionizing radiation than a comparable fragment of nDNA in animal cells [13,27-29]. Moreover, mtDNA fragments are detected in the cytosol of brain and spleen cells of γ-irradiated mice for a long period of time [29,30]. Fragments of mtDNA might get into the intercellular space and the blood flow after radiation damage of the organism [30-32]. The release of mtDNA from mitochondria can occur as the result of their damage and selective mitophagy [13-15]. This phenomenon can also be favored by postradiational activation of ROS production in the mitochondria of damaged cells. It is well known, that the increased level of ROS can persist in mitochondria for a period from several minutes to several days, depending on the type of cells and radiation dose [33,34] by mitochondrial nucleoid proteins [35] and can migrate to the nuclei or between different cells [36].
It is a well-known fact that, along with various types of damage, ionizing radiation induces DSB in DNA. A predominant way of repair of DSB in nDNA in the cells of higher eukaryotes is the NHEJ mechanism, without the involvement of extensive regions on the ends of breaks [37]. DSB repair in nDNA can occur by the NHEJ mechanism, with the "capture" of an mtDNA fragment into end break joining sites, as well as with the involvement of microhomologies located in terminal sequences [38]. It has been recently demonstrated that the integration of mtDNA into nDNA can predominantly take place in the accessible regions of chromatin [39,40]. It is possible that new numt-pseudogenes also arise due to damage of DNA by other exogenous and endogenous factors. The integration of new mtDNA fragments apparently not only changes the structure of the genome in the regions of integration, but also substantially influences the realization of genetic information and genome stability. It has been demonstrated that the heat stress causes the migration of organelles mtDNA in the nucleus and their integration into the nuclear genome in plants [41]. A number of publications indicate that the amount of numt-insertions in nDNA grows with organism age. This may be due to accumulation of nDNA damage and mtDNA destruction due to action of ROS which are formed in the process of cell metabolism [12,35]. Recently it has been shown that nDNA in the tissues of old rats has a several-times higher homology with mtDNA, as compared to that of young animals [42]. Later on, the group of Barja G [12] used PCR and hybridization to prove that the number of mtDNA insertions raises in nDNA of liver and in the brain of rats with aging. The authors assume that accumulation of mtDNA insertions in animal nDNA might have a significant impact on organism aging [12]. This observation is confirmed by studies conducted on yeast cells, which testified the increase in the amount of numt-pseudogenes in nDNA with chronological aging [43].
As far back as 30 years ago it was proposed that the integration of mtDNA into the nuclear genome might be essential not only for aging, but also for cancer etiology [44]. For instance, the nDNA of rat hepatoma induced by a chemical carcinogen was found to contain homology regions with mtDNA (genes ND6 and CO?1), which are not revealed in the nDNA of normal hepatocytes [45]. It was found that the mtDNA sequences (gene COXIII) are integrated into one of the two alleles of the nuclear gene c-myc in HeLa cells [46]. The nDNA of HeLa cells also contains nucleotide sequences of about 5 kb, corresponding to mtDNA genes: CO?1, ND4, ND4L and 12S rRNA [7]. The results of another study showed that the nDNA of tumor cells of mice and rats has more elements with mtDNA homology than the nDNA of their normal tissues [47]. The investigations of Liang [48] registered large fragments of mtDNA in the nDNA of human glioma, which are absent in the nDNA of neuroglial cells. According to another set of data, some mtDNA fragments are absent in the nDNA of normal cells but are integrated into the nDNA of cells of epithelial cervical cancer. There insertions of mtDNA strongly influence the expression of oncogene c-myc [11].
Additionally, there is evidence that there is a relation between some inherited diseases in humans and the appearance of mtDNA insertions in the encoded nuclear genes. It has been determined that six syndromes in humans are associated with the insertion of numt-pseudogenes in particular chromosomes [8-10,49-53]. Insertions of mtDNA in the coding gene of germ cells can be regarded as mutations, which can have serious consequences for embryogenesis and for the surviving of the offspring. It should be noted that the Pallister-Hall syndrome manifested as a functional disturbance of a key developmental gene as a result of a de novo insertion of an mtDNA fragment into the nuclear genome have been described earlier [8]. This mutation has appeared after insertion of an mtDNA fragment of 72 bp into the nucleotide sequence of the GLI3 gene in the 7th chromosome. The parents of the patient did not have such a mutation, and this mtDNA insertion could occur in the parental germ cell or at an early postzygotic stage of development of the baby. The family lived in a city with a high radiation level after the Chernobyl power plant accident [8].
Thus, the obtained experimental data suggest that ionizing radiation, which causes the destruction of mtDNA and DSB in nDNA and the activation of the repair system, can be regarded as a highly important factor capable of inducing de novo the generation of numt-pseudogenes.
Acknowledgements
This work was supported in part by the Russian Foundation of Fundamental Research (grant number 080400163). We thank Fomenko L. and Kuznetsova E. for technical assistance in the experiments.
Authors Contributions
S.A.A. and A.I.G. were involved in project conception and design. S.A.A. was involved in laboratory processing and data acquisition for DNA isolation, PCR Array and DNA-sequences. A.I.G. helped interpret data and drafted the final manuscript, which was revised and approved by authors.
References
-
Thorsness PE, Weber ER (1996) Escape and migration of nucleic acids between chloroplasts, mitochondria, and the nucleus. Int Rev Cytol 165: 207-234.
-
Bensasson D, Zhang D, Hartl DL, Hewitt GM (2001) Mitochondrial pseudogenes: evolution's misplaced witnesses. Trends Ecol Evol 16: 314-321.
-
Lopez JV, Yuhki N, Masuda R, Modi W, O'Brien SJ (1994) Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol 39: 174-190.
-
Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123-135.
-
Hazkani-Covo E, Zeller RM, Martin W (2010) Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet 6: e1000834.
-
Kleine T, Maier UG, Leister D (2009) DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol 60: 115-138.
-
Shay JW, Werbin H (1992) New evidence for the insertion of mitochondrial DNA into the human genome: significance for cancer and aging. Mutat Res 275: 227-235.
-
Turner C, Killoran C, Thomas NS, Rosenberg M, Chuzhanova NA, et al. (2003) Human genetic disease caused by de novo mitochondrial-nuclear DNA transfer. Hum Genet 112: 303-309.
-
Ricchetti M, Tekaia F, Dujon B (2004) Continued colonization of the human genome by mitochondrial DNA. PLoS Biol 2: E273.
-
Chen JM, Chuzhanova N, Stenson PD, Férec C, Cooper DN (2005) Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum Mutat 25: 207-221.
-
Chen D, Xue W, Xiang J (2008) The intra-nucleus integration of mitochondrial DNA (mtDNA)in cervical mucosa cells and its relation with c-myc expression. J Exp Clin Cancer Res 27: 36.
-
Caro P, Gómez J, Arduini A, González-Sánchez M, González-García M, et al. (2010) Mitochondrial DNA sequences are present inside nuclear DNA in rat tissues and increase with age. Mitochondrion 10: 479-486.
-
Gaziev AI (2013) [Pathways for maintenance of mitochondrial DNA integrity and mitochondrial functions in cells exposed to ionizing radiation]. Radiats Biol Radioecol 53: 117-136.
-
Kim I, Lemasters JJ (2011) Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid Redox Signal 14: 1919-1928.
-
Okamoto K, Kondo-Okamoto N (2012) Mitochondria and autophagy: critical interplay between the two homeostats. Biochim Biophys Acta 1820: 595-600.
-
Blanchard JL, Schmidt GW (1996) Mitochondrial DNA migration events in yeast and humans: integration by a common end-joining mechanism and alternative perspectives on nucleotide substitution patterns. Mol Biol Evol 13: 537-548.
-
Ricchetti M, Fairhead C, Dujon B (1999) Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature 402: 96-100.
-
Yu X, Gabriel A (1999) Patching broken chromosomes with extranuclear cellular DNA. Mol Cell 4: 873-881.
-
Decottignies A (2005) Capture of extranuclear DNA at fission yeast double-strand breaks. Genetics 171: 1535-1548.
-
Hazkani-Covo E, Covo S (2008) Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet 4: e1000237.
-
Gaziev AI, ShaĬkhaev GO (2010) [Nuclear mitochondrial pseudogenes]. Mol Biol (Mosk) 44: 405-417.
-
Pereira SL, Baker AJ (2004) Low number of mitochondrial pseudogenes in the chicken (Gallus gallus) nuclear genome: implications for molecular inference of population history and phylogenetics. BMC Evolut Biol 4: 17.
-
Fumihito A, Miyake T, Sumi S, Takada M, Ohno S, et al. (1994) One subspecies of the red junglefowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proc Natl Acad Sci U S A 91: 12505-12509.
-
Kost TA, Theodorakis N, Hughes SH (1983) The nucleotide sequence of the chick cytoplasmic beta-actin gene. Nucleic Acids Res 11: 8287-8301.
-
Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, et al. (2006) Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J 25: 5358-5371.
-
Desjardins P, Morais R (1990) Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol 212: 599-634.
-
May A, Bohr VA (2000) Gene-specific repair of gamma-ray-induced DNA strand breaks in colon cancer cells: no coupling to transcription and no removal from the mitochondrial genome. Biochem Biophys Res Commun 269: 433-437.
-
Malakhova L, Bezlepkin VG, Antipova V, Ushakova T, Fomenko L, et al. (2005) The increase in mitochondrial DNA copy number in the tissues of gamma-irradiated mice. Cell Mol Biol Lett 10: 721-732.
-
Patrushev M, Kasymov V, Patrusheva V, Ushakova T, Gogvadze V, et al. (2006) Release of mitochondrial DNA fragments from brain mitochondria of irradiated mice. Mitochondrion 6: 43-47.
-
Abdullaev SA, Antipova VN, Gaziev AI (2009) [Extracellular mutant mitochondrial DNA content is sharply elevated in the blood plasma of irradiated mice]. Mol Biol (Mosk) 43: 1063-1069.
-
Evdokimovsky EV, Ushakova TE, Kudriavtcev AA, Gaziev AI (2011) Alteration of mtDNA copy number, mitochondrial gene expression and extracellular DNA content in mice after irradiation at lethal dose. Radiat Environ Biophys 50: 181-188.
-
Zhang M, Zhang B, Guo Y, Zhang L, Yang S, et al. (2013) Alteration of circulating mitochondrial DNA concentration after irradiation. Adv Exp Med Biol 765: 371-377.
-
Kim GJ, Chandrasekaran K, Morgan WF (2006) Mitochondrial dysfunction, persistently elevated levels of reactive oxygen species and radiationinduced genomic instability: A review Mutagenesis 21: 361-367.
-
Azzam EI, Jay-Gerin JP, Pain D (2012) Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett 327: 48-60.
-
Gaziev AI, Abdullaev S, Podlutsky A (2014) Mitochondrial function and mitochondrial DNA maintenance with advancing age. Biogerontology 15: 417-438.
-
Spees JL, Olson SD, Whitney MJ, Prockop DJ (2006) Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci U S A 103: 1283-1288.
-
Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247-271.
-
Decottignies A (2013) Alternative end-joining mechanisms: a historical perspective. Front Genet 4: 48.
-
Tsuji J, Frith MC, Tomii K, Horton P (2012) Mammalian NUMT insertion is non-random. Nucleic Acids Res 40: 9073-9088.
-
Wang D, Timmis JN (2013) Cytoplasmic organelle DNA preferentially inserts into open chromatin. Genome Biol Evol 5: 1060-1064.
-
Wang D, Lloyd AH, Timmis JN (2012) Environmental stress increases the entry of cytoplasmic organellar DNA into the nucleus in plants. Proc Natl Acad Sci U S A 109: 2444-2448.
-
Mozzhukhina TG, Orlichenko LS, Litoshenko AIa (1994) [The presence of mtDNA-like sequences in the DNA of liver chromatin fractions from rats of different ages]. Izv Akad Nauk Ser Biol : 751-760.
-
Cheng X, Ivessa AS (2012) Accumulation of linear mitochondrial DNA fragments in the nucleus shortens the chronological life span of yeast. Eur J Cell Biol 91: 782-788.
-
Reid RA (1983) Can migratory DNA activate oncogenes? Trends in Biochem Sci 8: 189-226.
-
Corral M, Kitzis A, Baffet G, Paris B, Tichonicky L, et al. (1989) RNAs containing mitochondrial ND6 and COI sequences present an abnormal structure in chemically induced rat hepatomas. Nucleic Acids Res 17: 5191-5206.
-
Shay JW, Baba T, Zhan QM, Kamimura N, Cuthbert JA (1991) HeLaTG cells have mitochondrial DNA inserted into the c-myc oncogene. Oncogene 6: 1869-1874.
-
Hadler HI, Devadas K, Mahalingam R (1998) Selected nuclear LINE elements with mitochondrial-DNA-like inserts are more plentiful and mobile in tumor than in normal tissue of mouse and rat. J Cell Biochem 68: 100-109.
-
Liang BC (1996) Evidence for association of mitochondrial DNA sequence amplification and nuclear localization in human low-grade gliomas. Mutat Res 354: 27-33.
-
Willett-Brozick JE, Savul SA, Richey LE, Baysal BE (2001) Germ line insertion of mtDNA at the breakpoint junction of a reciprocal constitutional translocation. Hum Genet 109: 216-223.
-
Borensztajn K, Chafa O, Alhenc-Gelas M, Salha S, Reghis A, et al. (2002) Characterization of two novel splice site mutations in human factor VII gene causing severe plasma factor VII deficiency and bleeding diathesis. Br J Haematol 117: 168-171.
-
Goldin E, Stahl S, Cooney AM, Kaneski CR, Gupta S, et al. (2004) Transfer of a mitochondrial DNA fragment to MCOLN1 causes an inherited case of mucolipidosis IV. Hum Mutat 24: 460-465.
-
Ahmed ZM, Smith TN, Riazuddin S, Makishima T, Ghosh M, et al. (2002) Nonsyndromic recessive deafness DFNB18 and Usher syndrome type IC are allelic mutations of USHIC. Hum Genet 110: 527-531.
-
Millar DS, Tysoe C, Lazarou LP, Pilz DT, Mohammed S, et al. (2010) An isolated case of lissencephaly caused by the insertion of a mitochondrial genome-derived DNA sequence into the 5' untranslated region of the PAFAH1B1 (LIS1) gene. Human Genomics 4: 384 -393.