Genomic profiling (GP) of breast cancer (BC) tumors is a promising approach to identifying genetic mutations that aid patient management. Herewith, we present a single institutional experience of performing GP to assess its impact on patient outcomes. Tumor tissues of 56 patients were tested using the FoundationOne® next-generation sequencing platform. Patients median age was 54 years; 51 patients had metastatic BC, while 5 had an early-stage disease. The most frequently identified genes were TP53, PIK3, PTEN, and ESR1. Four and one patients had BRCA2 and BRCA1, respectively. Actionable mutations and potential matched treatment were identified in 44 (79%) and 27 (48%) patients. However, the GP findings guided further management decisions in only 18 (32%) patients. There was no difference in clinical benefit (complete response, partial response, and stable disease), median (95% CI) progression-free survival, and median overall survival (95% CI) between those whose management was GP-guided or unguided (35% vs. 48%; 24 [12-36.7] vs. 26.4 [13.7-39] months; 28.3 [12.4-44.3] vs. 30.5 [24.4-36.6] months, respectively). Despite a high prevalence of actionable mutations, GP-guided decision was possible in a few patients. The small sample size probably precluded the demonstration of favorable outcomes among those managed as guided by the GP findings.
Breast cancer, Metastatic, Genomic profiling, Survival
Breast cancer is a complex disease that stands as one of the most prevalent and heterogeneous malignancies that affects millions of women worldwide [1]. Over the past few decades, the oncology community has witnessed significant advancements in medical technology and understanding of the intricate molecular landscape of this disease.
Among these, somatic testing of breast cancer tumors is a promising approach to identifying genetic mutations that drive tumor growth and progression. This type of testing involves analyzing the DNA of cancer cells to identify mutations that are not present in normal cells. The elucidation of these molecular signatures not only aids in precise diagnostic classifications but also fuels tailored therapeutic approaches, fostering the era of personalized medicine.
Recently, the American Society of Clinical Oncology published its provisional clinical opinion and stated that “Patients with metastatic or advanced cancer should undergo genomic sequencing in a certified laboratory if the presence of one or more specific genomic alterations has regulatory approval as biomarkers to guide the use of or exclusion from certain treatments for their disease” [2]. In 2020 alone, 28 targeted therapies were approved by the FDA in patient populations defined by specific molecular biomarkers [3]. Nevertheless, most alterations detected by genomic sequencing are passengers with no impact on cancer development [4], and a smaller fraction of alterations are drivers [5]. A major challenge in precision oncology is determining whether a specific genomic alteration in a potentially targetable gene is a passenger, actionable, or nonactionable driver.
In this retrospective study, we intended to examine the impact of somatic genomic profiling (GP) testing on the characterization and management of patients with breast cancer in a single institution. By examining the key molecular alterations identified through these tests and their clinical implications, this research aims to underscore the significance of integrating genomic information into the comprehensive management of breast cancer.
Between January 2018 and July 2023, 56 consecutive women with breast cancer whose tumors were evaluated for GP were included in the current analysis, and their electron data were retrieved. Genomic profiling was requested mainly for patients with metastatic breast cancer and fewer patients with early-stage disease whose initial diagnosis is linked to a potential targetable mutation. The abstracted information included demographic data, clinicopathologic features, history of prior therapy, the next-generation sequencing (NGS) results, and their impact on management decisions, disease response, and survival outcome. The institutional review board approved the study and the consent form.
GP was performed on archival fixed formalin paraffin-embedded tissue from a tissue obtained from the primary tumor or a metastatic lesion if appropriate. We have used the commercially available NGS platform, FoundationOne ® (Foundation Medicine, Inc.). The NGS mutational analysis using the FoundationOne ® is based on a panel of 315 genes that drive cancer growth and introns from 28 genes involved in rearrangements [6]. The test results also included microsatellite stability/instability status, tumor mutational burden, and PD-L1 expression or amplification.
In a multidisciplinary molecular tumor board, the findings of the GP for each patient were discussed. Therapeutically actionable genes were defined as gene alterations that confer sensitivity or resistance to an available therapy [2], and treatment was considered "matched" if an agent(s) could target an aberration in a patient's GP or a functionally active protein expressed that may guide therapy decision. Otherwise, the treatment would be considered "unmatched." Upon obtaining the results, the treatment decision was either to recommend changing or initiating a different treatment as guided by the results (GP-guided decision), maintain the current treatment, or recommend changing or starting another treatment not driven by the results (GP-unguided decision).
Descriptive statistics of relevant data are provided. Objective response rate (ORR) was evaluated according to the Response Evaluation Criteria in Solid Tumors, version 1.1 [7], and the difference in ORR was compared using the odds ratio (OR) and its 95% confidence interval (CI). Progression-free survival (PFS) was computed between the implementation date of a management decision based on the GP's findings and the date of progression or death of any cause. Overall survival (OS) was calculated between the implementation date of a management decision based on the GP's results to the date of death of any cause or the date of the last contact. Survival functions were computed using the Kaplan-Meier method, and the survival difference between groups was compared using the log-rank test. All tests were two-sided at the 5% significance level. The statistical analyses were done with the SPSS statistical package (IBM SPSS Statistics for Windows, version 25.0., New York, USA).
Patients median age (95% CI) at the time of initial diagnosis and when GP was performed was 47 (42-53) and 54 (50-56) years, respectively. Table 1 shows patients and disease characteristics. While Table 1 shows that 18 patients had an early-stage disease at diagnosis, GP was performed upon the recurrence in 13 patients. GP was requested to guide further treatment of the remaining five patients who didn't experience recurrence, such as determining PD-L1 status or eligibility for adjuvant olaparib [8].
Table 1: Patient and disease characteristics. View Table 1
One or more mutational gene(s) were identified in every GP test, and Table 2 depicts the most common genes and other relevant findings. As shown, TP53, PIK3, PTEN, and ESR1 were the most frequently identified genes. Also noted that the positivity for BRCA2 was more frequent than that for BRCA1 (4 patients and one patient, respectively). Of note, no tumor showed positivity for CHEK2 or PALB mutation.
Table 2: Frequency of identified mutations and other findings. View Table 2
Analysis of the derived results for decision-making based on the consensus opinion at the multidisciplinary molecular tumor board is shown in Table 3. Actionable mutations were detected among 79% of patients. However, potential matched intervention was only feasible in 48% of cases. Table 3 also shows that the GP guided an immediate management decision upon obtaining the results for 32% of patients and provided an additional 9% guided decisions if a change in management, such as disease progression, is to be dictated in the future.
Table 3: Analysis of the genomic profiling results and management decision. View Table 3
This analysis is only restricted to the 51 patients after excluding the five patients with early-stage disease who didn't experience disease recurrence. Table 4 shows the ORR to the management decision as guided or unguided based on the GP findings. As shown, no significant difference existed between the adopted decisions in the ORR. Furthermore, there was no significant difference in the clinical benefit rates (combined CR, PR, and SD) between GP-guided or unguided management decisions (OR [95% CI] = 0.59 [0.17-2.05]; P = 0.40).
Table 4: Disease response to management decision. View Table 4
Again, this analysis is only restricted to the 51 patients after excluding the five patients with early-stage disease who didn't experience disease recurrence. The median follow-up (95% CI) from implementing the management decision based on the GP findings to the date of death or last contact, whichever comes first, was 28.3 (26.1-30.9) months. The median PFS (95% CI) for the 51 patients was 26.4 (16.6-36.3) months. Examining the median PFS based on the management decision is shown in Table 5, and there was no PFS difference, irrespective of the management decision (Figure 1).
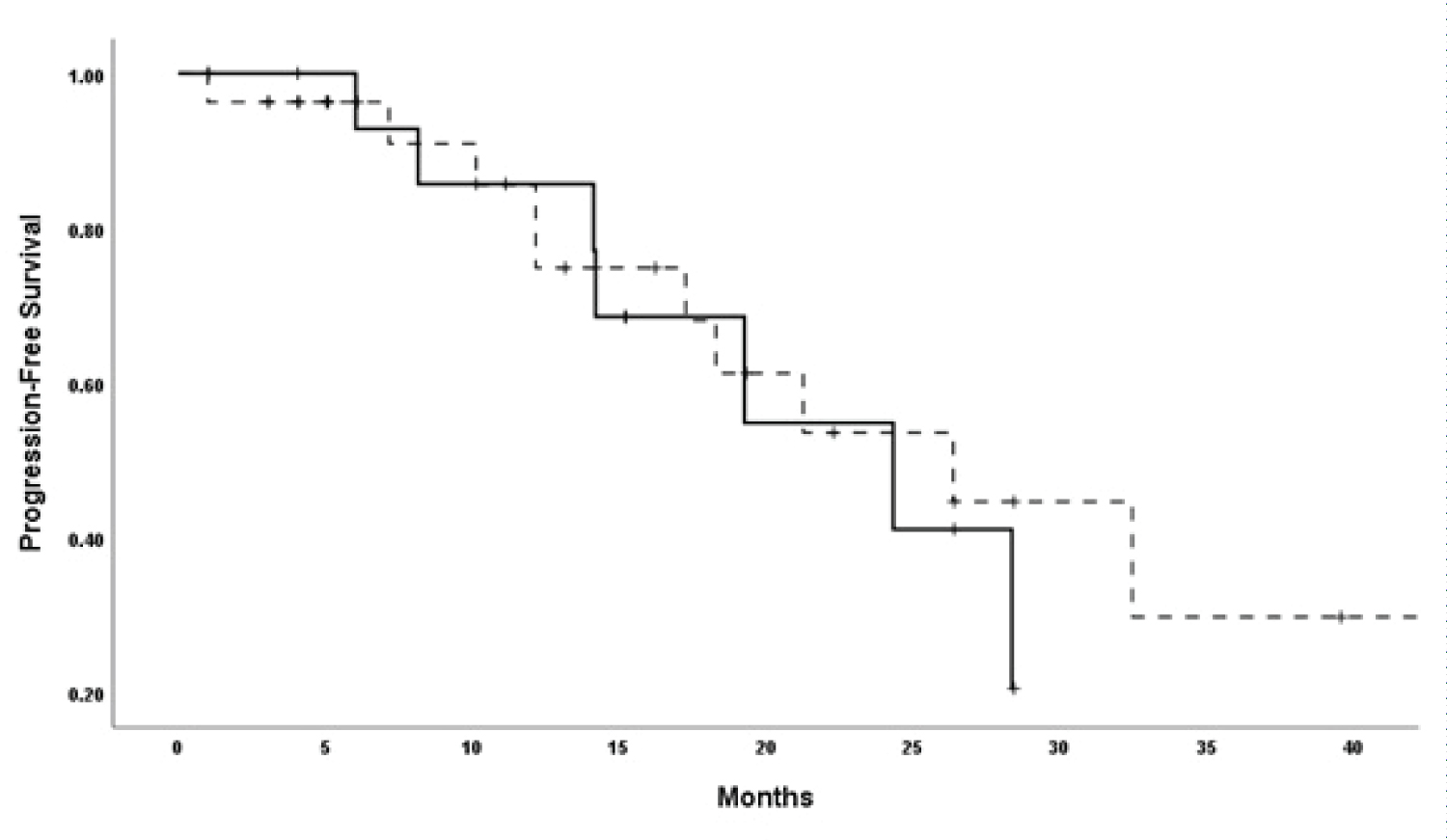 Figure 1: Progression-free survival of patients according to the management decision (solid line, genomic prefilling-guided decision, dashed-line, genomic profiling-unguided decision).
View Figure 1
Figure 1: Progression-free survival of patients according to the management decision (solid line, genomic prefilling-guided decision, dashed-line, genomic profiling-unguided decision).
View Figure 1
Table 5: Progression-free survival and overall survival according to the management decision guided or unguided by the genomic profiling findings. View Table 5
The median OS (95% CI) for the 51 patients was 29.5 (25.9-33.1) months. Examining the median OS based on the management decision is shown in Table 5. There was no OS difference between the groups (Figure 2).
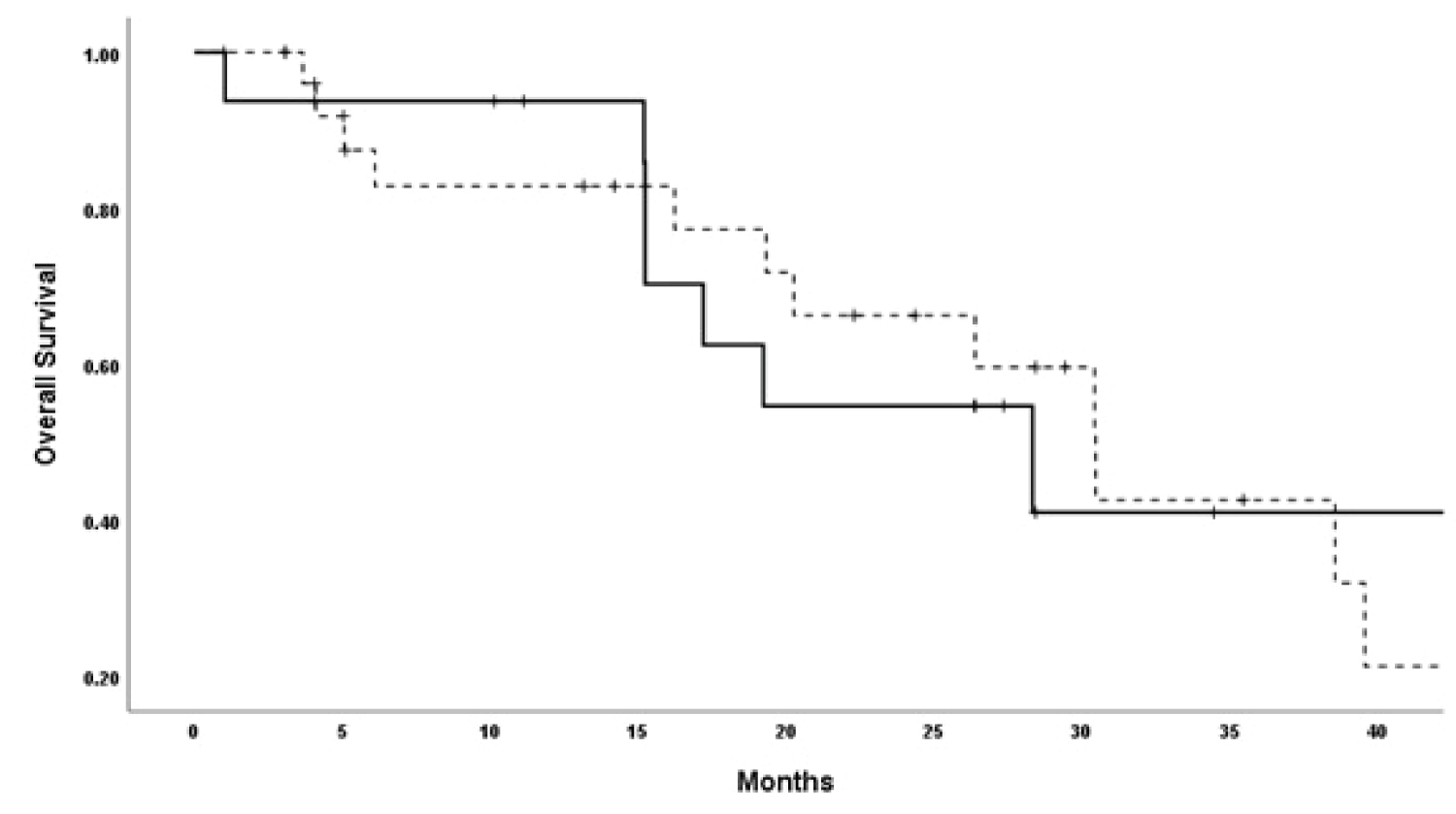 Figure 2: Overall survival patients according to the management decision (solid line, genomic prefilling-guided decision, dashed-line, genomic profiling-unguided decision).
View Figure 2
Figure 2: Overall survival patients according to the management decision (solid line, genomic prefilling-guided decision, dashed-line, genomic profiling-unguided decision).
View Figure 2
The utilization of GP in guiding therapeutic decisions has increasingly gained attention in oncology, promising a personalized approach towards cancer management. Notably, while GP was primarily requested upon recurrence, a subset of patients underwent GP without experiencing disease recurrence. This trend reflects the evolving paradigm of integrating genomic insights in advanced stages and potentially early-stage diseases for prognostic and therapeutic stratification.
Our findings revealed a diverse mutational landscape, with prevalent alterations observed in TP53, PIK3, PTEN, and ESR1 genes, indicating potential targets for therapeutic intervention. Interestingly, the prevalence of BRCA2 mutations exceeded that of BRCA1, suggesting distinct implications for targeted therapies such as PARP inhibitors.
The multidisciplinary molecular tumor board's consensus-based decision-making, guided by GP findings, played a pivotal role in shaping treatment strategies. It notably influenced immediate management decisions for a portion of patients, with additional guidance for potential future alterations in management. GP of some solid tumors and hematological malignancies has revolutionized precision oncology for those tumors and resulted in a significant survival advantage [9-11].
However, in this study, we could not demonstrate any significant difference in ORR, PFS, OS, and clinical benefit rates between GP-guided and unguided management decisions. Therefore, it may imply that while a GP informs decisions, it might not substantially alter short-term clinical outcomes or immediate treatment responses. The lack of significant survival differences in some clinical trials highlights the complexity of translating genomic insights into substantial survival benefits, necessitating a deeper understanding of the interplay between molecular alterations, treatment response, and long-term outcomes.
One such study focused on patients with high-risk early-stage breast cancer. The study found that although genomic profiling provided additional information about the risk of recurrence, it did not significantly improve survival outcomes [12]. In another study, the phase II SHIVA study, while not exclusively focused on breast cancer, the trial included breast cancer patients and investigated the clinical value of targeted therapies based on molecular profiling versus conventional therapy [13]. No PFS difference was demonstrated; however, in updated results, the authors reported that patients who crossed over to the experimental arm achieved a 30% improvement in PFS [14].
However, the current study has some limitations, such as its retrospective nature and relatively small sample size, warrant cautious interpretation of the findings. Future prospective studies with larger cohorts must validate these observations and elucidate the intricate dynamics between GP-informed decisions, treatment response, and patient outcomes. Moreover, there are many challenges faced by precision oncology that are yet to be overcome, such as the cost, accessibility, tissue availability, the need to improve methods to retrieve sufficient tumor material, intratumor heterogeneity, the identification and characterization of molecular subtypes of cancer and the mutations [15]. Furthermore, while an actionable alteration may be detected in approximately one-third of patients with solid tumors, only a smaller proportion of patients will be assigned [16].
• No funding was required.
• All authors contributed equally to the study.
• All authors declare that there was no conflict of interests.
• The study was approved by the Institutional Review board.
• The original data are available if requested.