There is increasing evidence that epigenetic factors may play a role in the pathogenesis of Systemic Lupus Erythematosus (SLE). Both global and gene specific methylation is known to occur in lupus patients, as well as, changes in histone acetylation status. Histone acetylation is associated with active chromatin or activation of genes, whereas histone deacetylase (HDAC) activity is associated with silencing of genes. Therefore, HDACs have been targeted as potential therapeutic targets for a number of diseases, including lupus. The purpose of this study was to determine histone deacetylase (HDAC) expression in patients who are diagnosed with SLE compared to age-matched healthy controls. Quantitative real-time PCR expression levels of HDAC 1, HDAC 2 and HDAC 7 were investigated in peripheral blood mononuclear cells of African American and European American women. Our results showed that HDAC 1 expression is significantly (p < 0.0039) elevated in lupus patients compared to controls. HDAC 2 expression is also increased in lupus patients (p < 0.0427). However, HDAC 7 showed no significant difference (p < 0.4644) in expression in our SLE patients compared to their controls. Those lupus patients with a SLE disease activity index (SLEDAI) of 4 or greater showed lower expression of HDAC 1 (p < 0.0026) compared to those with modest disease and a SLEDAI of less than 4. However, in those lupus patients with a SLEDAI of 4 or greater showed increased expression of HDAC2 (p < 0.053) when compared to those with a SLEDAI of less than 4. This observation was also noted in HDAC7. Increased expression in HDAC 1 and 2 has been associated with induced kidney injury and induction of proinflammatory cytokines.
Histone deacetylases, Systemic lupus erythematosus, Epigenetic regulation
The difficulties in designing an effective pharmacological therapy for Systemic Lupus Erythematosus (SLE) are due in part to the complexities of its pathophysiology. However, recently researchers have begun to look at the role chromatin modification plays in SLE. Chromatin modification is important in the regulation of genomic expression. One of the essential parts of chromatin structure are the histones. Histones are responsible for binding the nucleosome and provide the entry and exit sites to DNA. Like DNA, histones can also be subjected to epigenetic events. A group of enzymes that have shown to play a key role in histone modification are histone deacetylases (HDACs). HDAC enzymes work on the amino terminal tail of histones. They are able to regulate gene expression by modulating histone acetylation patterns [1]. There are 18 genes identified as HDACs. These 18 genes can further be grouped into four classes based on their structural functional capabilities. There has been increasing evidence that targeting certain classes of HDACs can provide therapeutic benefit to patients with SLE. For example, preclinical studies have shown that using the HDAC inhibitor (HDACi), Trichostatin A (TSA), can reduce anti-DNA autoantibody production [2]. In addition, suberorylanide hydroxamic acid (SAHA), a HDAC inhibitor, in combination with TSA can reduce IL-6, IL-12, IFN-γ, and IL-10 production leading to a decrease in inflammation response as well as decreases in proteinuria and glomerulonephritis in murine models [3,4]. Due to the effects of HDACi in their ability to reduce inflammation in murine models, researchers have postulated their use for patients to control inflammation such as those that occur with SLE [5-8]. However, studies regarding HDAC expression levels in patients with SLE are lacking. Here we compare expression levels of HDAC 1, HDAC 2 and HDAC 7 among patients with SLE to their age, sex, and ethnicity matched controls.
Patients participating in this study were part of the LUPUS study at the Brody School of Medicine at East Carolina University. This study consisted of a total of 224 participants. The participants representing this study were ninety-six women diagnosed as having SLE based on SLE disease activity index (SLEDAI) scores and by anti-dsDNA antibody analysis and ninety-one controls that were age, sex, and ethnicity matched. No males were analyzed in this present study. Informed consent was obtained from all of our participants for blood samples to conduct our analysis. Also, this project was granted IRB approval from both The US Food and Drug Administration and from East Carolina Brody School of Medicine.
For our analysis, blood samples were collected by venipuncture of the antecubital vein between 9:00 AM-12:00 PM. In order to maintain a similar circadian pattern between our participants with SLE and their matched control participants, collections were conducted at the same time of day and the same day of the week. Peripheral blood mononuclear cells were isolated from whole blood using PAXgene RNA Blood tubes at East Carolina University in Greenville, NC, placed in dry ice and stored at -80 ℃ until shipped to the National Center for Toxicological Research for analysis.
RNA was extracted from peripheral blood mononuclear cells (PBMC) using a PAXgene RNA kit (QIAGEN, Valencia, CA). After extraction, all samples were tested for RNA integrity and concentration using a Bio-Rad Experion Automated Electrophoresis System (BIO-RAD, Hercules, CA). cDNA was created from RNA extractions using a Clontech Advantage® RT-for-PCR Kit (Clontech, Mountain View, CA).
HDAC 1, HDAC 2, and HDAC 7 mRNA expression was conducted using a Bio-Rad IQ5 quantitative Real Time Polymerase Chain Reaction Detection System (BIO-RAD, Hercules, CA). GAPDH was used as a housekeeping gene and as an endogenous control. qRT-PCR conditions were as follows: 50 ℃ for 2 minutes, 95 ℃ for 10 minutes (95 ℃ for 10 seconds, 56 ℃ for 45 seconds, 72 ℃ for 30 seconds) × 30 cycles. Relative quantitation's of HDAC 1, HDAC 2 and HDAC 7 mRNA expressions were normalized to GADPH and fold changes were calculated using a 2-ΛΛCT method. Primers utilized for both the histone deacetylases and GAPDH are listed in Table 1.
Table 1: Primers used for Quantitative Real Time Polymerase Chain Reaction. View Table 1
Statistical analyses were performed using GraphPad Prism Software Version 6.0 (San Diego, CA). A t-test were used for statistically significance. P < 0.05 was determined to be significant.
In Figure 1 we compare mRNA expression of HDAC 1 from normal controls and lupus patients. Individual differences were noted among the patients. In addition, statistically significant differences in HDAC1 mRNA expression were observed between lupus compared to non-lupus. Furthermore, HDAC 1 mRNA expression levels were significantly higher (p < 0.0039) in patients with SLE compared to controls. In Figure 2, we determined the mRNA expression levels of HDAC 2 from normal and lupus patients. Our analysis indicated that HDAC 2 expression levels was significantly higher (p < 0.0427) in our SLE patients compared to our controls. However, HDAC 7 mRNA expression levels among SLE and control patients were not significantly different (Figure 3 and Figure 4) (p < 0.4644). Our results provide evidence that histone deacetylases may be involved in the pathogenesis of SLE and Class I HDACs should be further investigated as potential therapeutic targets. Furthermore, these results demonstrated that an increase in HDAC 2 (p < 0.053) and 7 (p < 0.0259) expression were observed more in lupus patients with a SLE disease activity index (SLEDAI) of 4 or higher, when compared to patients with more modest disease, Figure 5 and Figure 6. However, a decrease in expression of HDAC 1 (p < 0.0026) was noted in lupus patients with higher SLEDAIs.
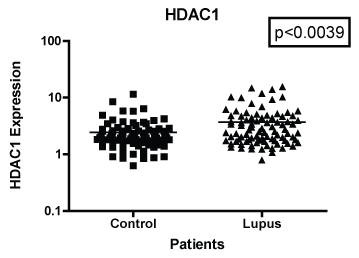 Figure 1: Note: HDAC 1 mRNA expression among women with SLE compared to age-matched controls. GAPDH was used as the housekeeping gene for comparison. The p-value was (p < 0.0039) which indicated a significant difference between the two populations.
View Figure 1
Figure 1: Note: HDAC 1 mRNA expression among women with SLE compared to age-matched controls. GAPDH was used as the housekeeping gene for comparison. The p-value was (p < 0.0039) which indicated a significant difference between the two populations.
View Figure 1
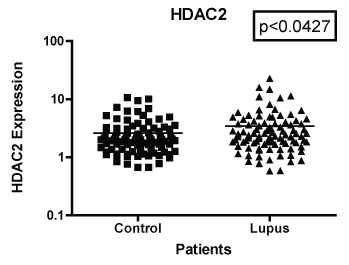 Figure 2: Note: HDAC 2 mRNA expression both African American and European American women with SLE vs. aged, ethnicity and sex matched Controls. GAPDH was used as the housekeeping gene for comparison. The p-value was (p < 0.0427) which indicated there were significant difference between the two populations.
View Figure 2
Figure 2: Note: HDAC 2 mRNA expression both African American and European American women with SLE vs. aged, ethnicity and sex matched Controls. GAPDH was used as the housekeeping gene for comparison. The p-value was (p < 0.0427) which indicated there were significant difference between the two populations.
View Figure 2
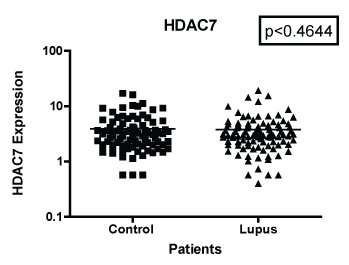 Figure 3: Note: HDAC 7 mRNA expression among women with SLE vs. matched Controls. GAPDH was used as the housekeeping gene for comparison. The p-value was (p < 0.4644) which indicated was no significant differences between the two populations.
View Figure 3
Figure 3: Note: HDAC 7 mRNA expression among women with SLE vs. matched Controls. GAPDH was used as the housekeeping gene for comparison. The p-value was (p < 0.4644) which indicated was no significant differences between the two populations.
View Figure 3
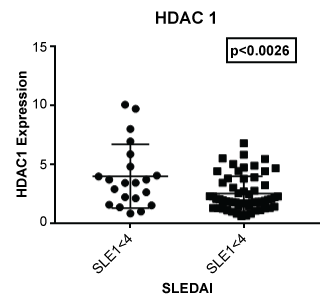 Figure 4: Expression of HDAC 1 in lupus patients with a SLE disease activity index (SLEDAI) of less than 4 (SLEI < 4) and 4 or greater (SLEI > 4). Those with more severe disease had a lower expression of HDAC1 (p < 0.0026) compared to those with modest disease.
View Figure 4
Figure 4: Expression of HDAC 1 in lupus patients with a SLE disease activity index (SLEDAI) of less than 4 (SLEI < 4) and 4 or greater (SLEI > 4). Those with more severe disease had a lower expression of HDAC1 (p < 0.0026) compared to those with modest disease.
View Figure 4
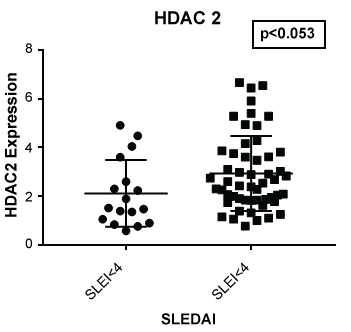 Figure 5: Expression of HDAC 2 in lupus patients with a SLE disease activity index (SLEDAI) of less than 4 (SLEI < 4) and 4 or greater (SLEI > 4). Those with more severe disease had a higher expression of HDAC2 (p < 0.053) compared to those with modest disease or with a SLEDAI of less than 4.
View Figure 5
Figure 5: Expression of HDAC 2 in lupus patients with a SLE disease activity index (SLEDAI) of less than 4 (SLEI < 4) and 4 or greater (SLEI > 4). Those with more severe disease had a higher expression of HDAC2 (p < 0.053) compared to those with modest disease or with a SLEDAI of less than 4.
View Figure 5
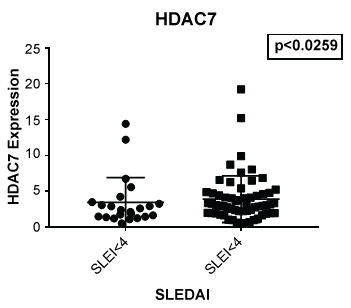 Figure 6: Although no significant difference was noted between lupus and non-lupus in expression of HDAC 7, lupus patients with a SLEDAI of 4 or greater showed an increased expression of HDAC7 (p < 0.0259) when compared to those with a SLEDAI of less than 4.
View Figure 6
Figure 6: Although no significant difference was noted between lupus and non-lupus in expression of HDAC 7, lupus patients with a SLEDAI of 4 or greater showed an increased expression of HDAC7 (p < 0.0259) when compared to those with a SLEDAI of less than 4.
View Figure 6
Histone deacetylases (HDACs) are critical for the maintenance of gene and chromosome silencing. Furthermore, HDACs assist in chromatin modification and transcriptional regulation of an organism's genome. In the preset study, we were interested in determining if HDAC expression was altered in Lupus patients as compared to non-Lupus patients. Our results demonstrated that the mRNA expression levels of Class I HDACs among SLE patients compared to controls were significantly different. Specifically, HDAC 1 and 2 were significantly up-regulated in SLE patients compared to controls. However, HDAC 7, which is a member of the class II HDACs, did not show a significant difference in expression level between SLE and controls patients. Of the histone deacetylases studied, HDAC 1 had significantly higher mRNA expression than HDAC 2 and 7. There is increasing evidence that over expression of HDAC 1 is linked to various malignancies such as cancer and kidney damage [9]. In addition, studies have shown that an increase in HDAC 1 expression is linked to decrease survival rates [10,11]. This suggests that HDAC 1 expression could be a useful biomarker for disease progression in SLE patients. Furthermore, HDAC 1 may be a potential therapeutic target for pharmacological design. On the other hand, HDAC 2 showed a significant difference in expression between SLE and control patients however, this difference was not as significant as HDAC 1 expression. Researchers have provided evidence that HDAC 2 expression levels can be linked as a possible biomarker for survival; especially in cases of oral cancer [12]. However, in cancer models, HDAC 2 has been shown to play an anti-apoptotic role [13]. With one of the hallmarks of SLE being apoptotic complications, HDAC 2 targeting could prove a potential avenue for therapeutic analysis. This is further underscored by the fact that this study demonstrated an increase in expression of HDAC 2 in the SLE population. This suggests that HDAC 2 should be further investigated in SLE.
Parts of this work was funded by a grant to Dr. Lyn-Cook from the FDA Office of Women's Health.
The findings and results reported in this manuscript are those of the authors and do not necessary represent the views of the US Food and Drug Administration.