Invasive listeriosis is a rare but life-threatening infection caused by Listeria monocytogenes. Human invasive listeriosis is considered a foodborne disease and contaminated ready-to-eat foods represent high-risk sources of human listeriosis [1]. Listeriosis generally affects immunocompromised individuals, pregnant women and newborns. In this risk group, listeriosis has a high lethality rate of 20 to 30% [2]. The infection is characterized by various clinical conditions such as spontaneous abortions, meningoencephalitis, septicaemia, gastroenteritis, and serious infections to the newborns [3]. Pregnant women are 15 to 18 times more likely to develop listeriosis after consumption of food contaminated with L. monocytogenes as opposed to the general population [4]. They tend to have mild clinical symptoms or be asymptomatic but the infection can have severe outcome for the fetus or newborn infant including miscarriage, stillbirth, neonatal sepsis and/or meningitis [5].
We performed a retrospective study in order to report culture-confirmed cases of pregnancy-related listeriosis occurring in Argentina from 1986 to 2016, to describe serotype distribution and to assess the genetic diversity of L. monocytogenes isolates.
One case of pregnancy-related listeriosis was defined as a pregnant woman or newborn infant aged < 28 days from whom L. monocytogenes has been isolated from a sterile site such as the blood, placenta or cerebrospinal fluid. Based on epidemiological and clinical data available, cases were classified as: Maternal infection with ongoing pregnancy, fetal loss and live born neonatal listeriosis. Fetal loss was defined as the death of the fetus that occurred prior to the complete expulsion or removal from the body of the mother, whatever the gestational age, according to WHO-PAHO definition [6].
From a Culture Collection of 334 human listeriosis isolates, a set of 111 isolates from pregnancy-associated listeriosis cases collected by the National Reference Laboratory (NRL) from 1986 to 2016 were studied. The strains had been isolated and identified by standard methods in the routine clinical microbiology laboratories of the healthcare facilities from 11 provinces of Argentina.
Species identification was confirmed using morphological and biochemical tests [7]. Serotyping was carried out by a multiplex PCR which is based on the amplification of the following target genes: prs, lmo0737, lmo1118, open reading frame 2110 (ORF2110), ORF2819, and prfA as described by Doumith, et al. [8]. This PCR assay classifies strains into five distinct molecular serogroups: IIa (serotypes 1/2a, 3a), IIb (serotypes 1/2b, 3b, 7), IIc (serotypes 1/2c, 3c), IVa (serotypes 4a, 4c), and IVb (serotypes 4b, 4ab, 4d, 4e) and the variant profile of molecular serogroup IVb (IVb-v1) described in 2011 [9]. In a previous study we compared this PCR with the conventional agglutination method in a collection of more than 300 isolates of L. monocytogenes recovered from human listeriosis cases, food and environment. We found 98% concordance between both serotyping methods [10], therefore, we implemented multiplex PCR as an alternative to laborious and expensive classical serotyping. The control strains used for the PCR reactions were L. monocytogenes 1/2a CCBE105/04-380, L. monocytogenes 1/2b CCBE105/04-582, L. monocytogenes 1/2c CCBE105/04-754, L. monocytogenes 3a CCBE105/04-436, L. monocytogenes 3c CCBE105/04-412, L. monocytogenes 4a CCBE105/04-988, L. monocytogenes 4b CCBE105/04-868 and L. innocua CCBE105/04-90. These strains belong to the Special Bacteriology Culture Collection (CCBE) and were serotyped with conventional somatic and flagellar antigen serum agglutination using both slides and tubes (Denka Seiken Co, LTD, Japan).
From 2007, L. monocytogenes isolates from pregnancy-associated listeriosis cases (n = 21) were routinely subtyped by DNA macro restriction and pulsed-field gel electrophoresis (PFGE) as a means to survey for outbreaks, according to Center for Disease Control and Prevention PulseNet standardized procedure, using the restriction enzymes AscI and ApaI [11]. A set of 23 isolates received at the NRL from 1996 through 2005 was selected and PFGE was performed retrospectively as part of this study. Salmonella enterica serovar Braenderup, strain H9812, restricted with XbaI was used for molecular weight determinations in all PFGE gels. The cluster analysis was made by a similarity matrix calculation using the Dice coefficient and dendrograms were generated with BioNumerics 5.10 software (Applied Maths, St-Martens-Latern, Belgium) using the unweighted pair group method with arithmetic mean (UPGMA) with tolerance set to 1.5%. A concatenated analysis of both similarity matrixes of ApaI and AscI fingerprints was performed resulting in a cluster analysis characterizing all strains on the average similarity using both methods. Isolates were considered having the same pulsotype if they had identical band patterns (no single-band differences) with both enzymes. A cluster was defined when 2 or more isolates showed indistinguishable pulsotype with both enzymes.
The percentage of pregnancy-associated listeriosis cases isolated per decade is shown in Figure 1.
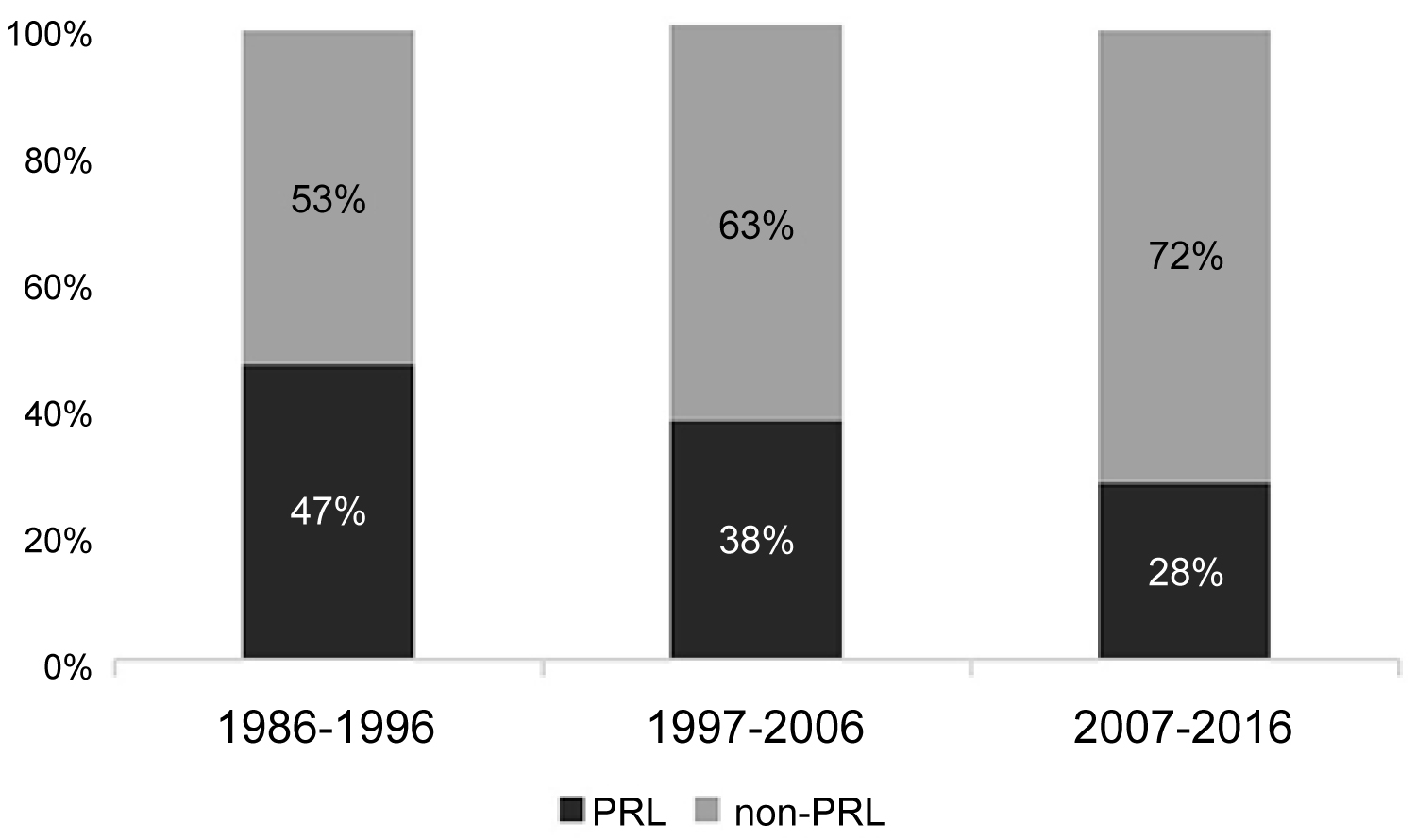 Figure 1: Percentage of culture confirmed cases of pregnancy related listeriosis per decade during the period 1986-2016. (PRL: pregnancy related listeriosis cases; non-PRL: listeriosis cases non associated to pregnancy).
View Figure 1
Figure 1: Percentage of culture confirmed cases of pregnancy related listeriosis per decade during the period 1986-2016. (PRL: pregnancy related listeriosis cases; non-PRL: listeriosis cases non associated to pregnancy).
View Figure 1
Cases were classified as: Maternal infection with ongoing pregnancy (12%); foetal loss (7%) and live born neonatal listeriosis (81%). Information about gestational age from foetal loss cases (n = 8) was available for 6 women (third trimester of pregnancy) and one woman (second trimester). Isolates from live born neonatal listeriosis were recovered from blood (72%), cerebrospinal fluid (17%) and both blood and cerebrospinal fluid (11%). All the strains isolated from maternal infection with ongoing pregnancy were recovered from blood cultures of the pregnant women, no miscarriage was reported. Serogroup IVb was prevalent (66%), followed by serogroup IIb (31%). Two isolates were assigned to serogroup IIa (2%) and one isolate to serogroup IIc (1%).
Molecular characterization of the 44 isolates of L. monocytogenes revealed 25 different PFGE types with AscI, whereas 26 patterns were identified using ApaI. One strain was no typeable by PFGE. Combining patterns from both enzymes resulted in 33 distinguishable types. As shown in Figure 2. Six clusters were identified which grouped isolates with indistinguishable fingerprint when analysed with both enzymes. They were arbitrarily named cluster A (4 isolates), cluster B (3 isolates) and clusters C, D, E and F with 2 isolates each of them. All the isolates grouped in clusters belonged to serogroup IVb. Since the NRL implemented PFGE as a means to survey for outbreaks, one cluster (cluster D) was detected and epidemiological data suggested that it might correspond to a probable nosocomial transmission involving 2 isolates of two newborns born consecutively in the same hospital in 2011. The NRL reported immediately to the Hospital and epidemiological investigations found no evidence of a food-borne or environmental source. The timing of onset of these infections within the same unit suggested cross-infection of L. monocytogenes between the two neonates. Blood culture and vaginal discharge from one mother were analysed and yielded negative for L. monocytogenes. Unfortunately, no clinical specimens were available from the other mother. Clusters A to C, E and F were identified as part of the retrospective analysis carried out for this study, therefore, epidemiological investigation could not be performed. Nevertheless, these clusters included identical bacterial clones isolated in a short period of time, pointing to possible common sources of infection.
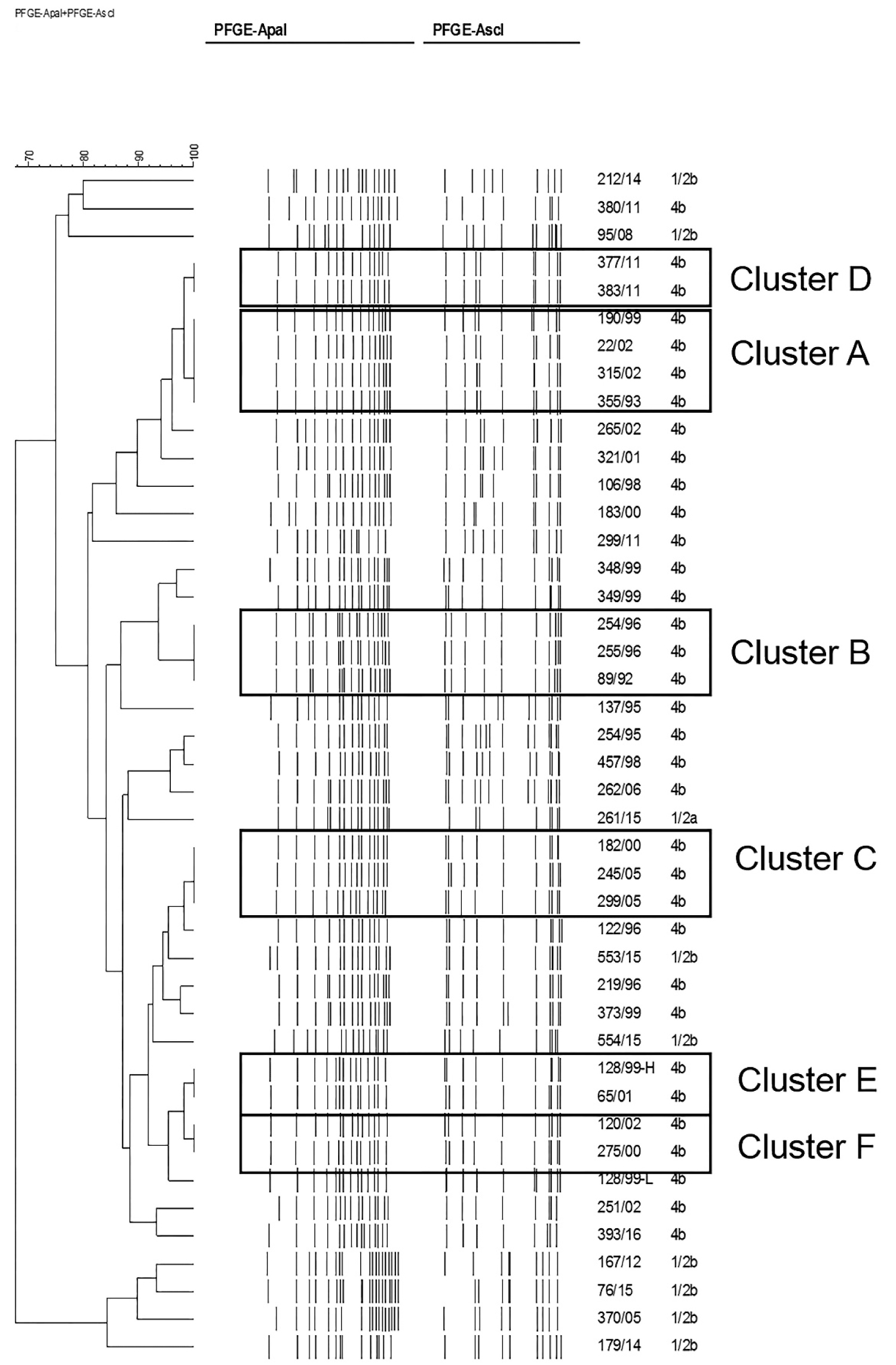 Figure 2: Genetic relationships of Listeria monocytogenes isolates from pregnancy related cases, based upon comparison of pulsed-field gel electrophoresis profiles obtained with the restriction enzymes ApaI and AscI. The dendrogram was produced with a Dice similarity coefficient matrix, using the unweighted pair group method with arithmetic mean (UPGMA). Tolerance and optimization values were set to 1.5%. Clusters are arbitrarily designated A to F.
View Figure 2
Figure 2: Genetic relationships of Listeria monocytogenes isolates from pregnancy related cases, based upon comparison of pulsed-field gel electrophoresis profiles obtained with the restriction enzymes ApaI and AscI. The dendrogram was produced with a Dice similarity coefficient matrix, using the unweighted pair group method with arithmetic mean (UPGMA). Tolerance and optimization values were set to 1.5%. Clusters are arbitrarily designated A to F.
View Figure 2
In recent years, epidemiology of pregnancy-related listeriosis has been investigated and well documented in several developed countries, such as England and Wales [12], France [13], United States [14], Israel [5] and Italy [15]. Listeriosis has been largely underreported in developing countries. There are few data about listeriosis collected in South America. Sporadic cases of listeriosis has been described in the region during the last 30 years [16-19] but the first listeriosis outbreaks with a clear epidemiological association between the consumption of a particular type of food were reported in Chile [17]. One outbreak occurred in 2008 with 165 reported cases and 14 deaths and was associated with the consumption of two types of soft cheese, a second outbreak in 2009 with 73 reported cases and 17 deaths was associated to consumption of sausage [17]. This is the first report describing the burden and trend of pregnancy associated listeriosis in Argentina and assessing the genetic diversity of L. monocytogenes isolates. Figure 2 showed a decrease in the number of cases of pregnancy-associated listeriosis during the last decade in Argentina. This observation is coincident with the improvement of the National Laboratory Network Systems in the country and the implementation of training and educational programs targeting biomedical community to improve prevention measures, early diagnosis and surveillance of the disease. In Argentina an effort to strengthen quantitative microbiological criteria, with a zero tolerance policy for foods that support the multiplication of bacteria has been implemented during the late 90s and recently regulations for L. monocytogenes aiming to the absence of this pathogen in 25 g of sample has been extended to ready-to-eat food. During the last decade, the improvement in control measures implemented at the local food production level might have impacted in the decrease of the number of cases. The retrospective molecular subtyping analysis showed clusters with isolates temporarily related which could represent under detected outbreaks.
The report of cases of listeriosis is not mandatory in Argentina. Argentinean statistics on human listeriosis has been based on a laboratory surveillance of the strains voluntary submitted to the National Reference Laboratory [20]. Underreporting might exist and routine surveillance of both cases and food is therefore incomplete. This study reports that 33% (111/334) of the identified listeriosis cases in Argentina were pregnancy-related and such frequency is similar to the rates reported in other countries such as Spain 32.5% and Israel 35%) [5,21]. Effective surveillance of listeriosis requires information about types of food consumed by the patients in order to assess risk and improve the awareness of dietary recommendations in susceptible population. Better centralized collection and subtyping of clinical and food isolates of L. monocytogenes would improve listeriosis monitoring, making it possible to detect outbreaks in real time and trace the sources of infection in order to enable rapid recalls of potentially contaminated products.
We thank Mr. Gastón Dangiolo and Mrs. Laura Polito for technical assistance.
None.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.