Methicillin resistant Staphylococcus aureus (MRSA) as a major cause of infections in hospital and community settings is a global health concern. The purpose of this study was to determine the antimicrobial susceptibility and the molecular characteristics of MRSA strains causing community-acquired (CA) and hospital-acquired (HA) infections in Tunisia.
A total of 135 non-duplicate MRSA strains were consecutively collected from five Tunisian hospitals. Antimicrobial susceptibility was done by disc diffusion method and by MIC. The presence of pvl (Panton Valentine Leukocidin) and tst-1 (toxic shock syndrome toxin 1) Genes were determined by PCR method. Strains were typed by agr, SCCmec typing, PFGE and spa typing.
Forty-nine strains (36.3%) were CA. HA strains showed significantly higher rates of resistance than the CA strains. One HA strain was resistant to teicoplanin (MIC = 4 µgml-1). The pvl gene was detected in 83.7% and 32.6% of CA and HA strains, respectively. Only eight strains were tst-1 positive. PFGE revealed 61 pulsotypes among HA strains and 20 pulsotypes among CA strains. Twenty-four spa types were identified. spa type t044 was the most common, representing 69.4% and 25.6% among CA and HA strains respectively. Most of t044 strains was pvl-positive, harbored agr3 and SCCmec IV and were resistant to kanamycin, tetracycline and fusidic acid. t037, agr1 and SCCmec III was the most prevalent among HA-MRSA.
Genetically diverse MRSA strains were circulating in our hospitals with relatively high prevalence of spa type t044 and t037. Regular surveillance studies on MRSA are needed to monitor the evolution of antimicrobial susceptibility, to better elucidate the distribution of existing MRSA clones and to detect the emergence of new MRSA clones.
Methicillin-Resistant Staphylococcus aureus, Epidemiology, pvl, agr, SCCmec, spa type
Over the last six decades, methicillin-resistant Staphylococcus aureus (MRSA) spread over the whole world and has become a global public health threat. Primarily MRSA was restricted to hospitals (hospital-acquired MRSA, HA-MRSA) and generally affect patients with predisposing risk factors such as recent hospitalisation, presence of indwelling catheters, admission to intensive care unit, exposure to a patient who is colonised or infected with MRSA, prolonged antibacterial therapy and surgery. For the last three decades, MRSA could also be found outside the hospital, in the community (community-acquired MRSA, CA-MRSA) and generally affect healthy and younger people without such the aforementioned risk factors [1,2]. Moreover, HA-MRSA and CA-MRSA belong to distinct genetic lineages. HA-MRSA strains are mostly multidrug resistant and carry the larger staphylococcal cassette chromosome mec (SCCmec) types I,II,III, however CA-MRSA strains frequently carry smaller SCCmec elements, usually type IV and V, and are resistant to fewer classes of antimicrobials. Also, CA-MRSA strains are strongly associated with virulence factors such as Panton Valentine leukocidin (PVL) which is thought to contribute to their pathogenicity [1-3]. Importantly, in recent years, the distinction between HA-MRSA and CA-MRSA has become increasingly blurred. In fact, CA-MRSA strains have been increasingly identified as a cause of HA-infections. On the other hand, HA-clones have been described to cause CA-infections suggesting that certain clones have the ability to cross barriers between hospitals and the community [3-7]. Recently, another development was the emergence of so-called livestock-associated MRSA (LA-MRSA) linked with mainly pig farming and is therefore found in people with contact to animals. These strains mainly belonged to the sequence type (ST) 398 [8]. In 2011, a novel mecA homologue, mecLGA251, named mecC located in a new SCCmec cassette designed SCCmec XI, has been reported from humans and animals in the Germany [9], UK and Denmark [10], Spain [11] and Austria [12]. These isolates mainly belonged to ST 130 [9-12].
Fortunately, whereas HA-MRSA isolates are generally multidrug resistant, CA and livestock-associated -MRSA tend to be resistant to fewer classes of antibiotics. The global emergence and spread of multidrug resistant MRSA limits the effectiveness of therapeutic options. Vancomycin has been the mainstay of treatment for MRSA infections and the emergence of resistance is rare and worrying [13,14].
Few studies have focused on molecular epidemiology of MRSA in Tunisia, so we conducted this retrospective multicenter study to investigate the molecular characteristics and the resistance profiles of MRSA causing community-acquired and hospital-acquired infections in Tunisia.
From March 2011 to March 2012, 135 clinical non-duplicate consecutive MRSA strains recovered from patients in five Tunisian university hospitals were studied: Habib Bourguiba hospital of Sfax which drains the south of Tunisia and it is located 270 km from the capital, Tunis (49 strains), Fattouma Bourguiba hospital of Monastir located in the center, 160 km from Tunis (25 strains), Institute Kassab of Tunis (26 strains), Charles Nicolle hospital of Tunis (22 strains) and military hospital of Tunis (13 strains). For each strain, demographic and clinical informations were recorded. CA-MRSA infection was defined as a positive culture of MRSA in patients with no history of hospitalization, surgery or outpatient care, or alternatively, if the signs of infection were present on admission. HA-MRSA infection was assigned when the strain was obtained at least 78h after hospitalization and when the infection was not the reason for admission [1].
The strains were identified by conventional methods: Gram-positive cocci, catalase positive, mannitol fermenting, DNase-positive and producing clumping factor (staphyslide test, bioMérieux).
The antibiotic susceptibilities of the strains were performed by a disc diffusion method on Mueller-Hinton agar (BioRad Laboratories, France) according to the CA-SFM criteria (http://www.sfm.asso.fr).
Methicillin resistance was confirmed by the detection of mecA gene by PCR as described elsewhere [15]. S. aureus ATCC 43300 was used as positive control (MRSA).
Minimal inhibitory concentrations (MICs) of vancomycin, teicoplanin, linezolid and tigecycline were determined by the broth microdilution method and were interpreted according to the CA-SFM criteria. Detection of heteroresistant vancomycin intermediate S. aureus strains (h-VISA) was performed by the Macromethod Etest on BHI agar for all of MRSA strains with a MIC ≥ 1 µgml-1 for vancomycin or teicoplanin, as described previously [16]. The strains h-VISA-Mu3 and Mu50 and the vancomycin-susceptible S. aureus ATCC 29213 were tested in parallel as positive and negative controls, respectively.
Genomic DNA used for polymerase chain reaction (PCR) was extracted using Instagène Matrix (BioRad) according to the manufacturer's instructions.
All strains were screened for the Panton Valentine Leukocidin, pvl (lukS-PV,lukF-PV) and toxic shock syndrome toxin 1, tst-1 genes by PCR simplex, as described previously [17,18]. S. aureus ATCC 49775 and S19 were used as controls for detection of pvl and tst-1 genes respectively.
The identification of agr groups was performed by multiplex PCR amplification of the hypervariable domain of the agr locus using primers specific for each of the four major specificity groups (forward primer, pan agr; and four reverse primers) [19]. S. aureus RN 6390 (agr group 1), RN 6607 (agr group 2), RN 8462/5 (agr group 3), and RN4850 (agr group 4) were used as controls for agr group identification.
The SCCmec types (I-IV) were detected by using the method described by Oliveira and de Lencastre [20]. Strains NCTC10442, N315, 85/2082, 4744, and WIS harboring, respectively, SCCmec type I-V were used as controls.
PFGE of chromosomal DNA after smaI macrorestriction was performed using the GenePath system (Bio-Rad, France) as described previously [21]. PFGE patterns were compared by using Finger Printing 2 Software (Biorad). Similarity matrice and dendrograms were obtained using arithmetic average (UPGMA). Similarity coefficients were calculated according to the dice method. Strains clustering above 80% similarity were considered the same clone.
spa typing was performed as previously described [22]. The X region of the spa gene was amplified by PCR. Purified spa PCR products were sequenced. spa types were determined with the Ridom Staph Type Software (Ridom, Gmbh Würzburg, Germany) which automatically detects spa repeats and assigns a spa type according to http://spaserver.ridom.de/.
X2 test was used to analyse the qualitative variables. A p value of < 0.05 was considered statistically significant.
Among the 135 strains studied, 49 (36.3%) were CA and 86 (63.7%) were HA. Clinical characteristics of patients with MRSA infection according to the origin of infection are summarized in Table 1.
Table 1: Demographic and clinical characteristics of patients with MRSA infection according to the origin of infection. View Table 1
The rates of resistance to antibiotics were low in CA-MRSA strains except for kanamycin (83.7%), tetracycline (75.5%), and fusidic acid (75.5%) (Table 2).
Table 2: Resistance rates of community-acquired (CA) and Hospital-acquired (HA) MRSA in Tunisia. View Table 2
Vancomycin, teicoplanin, linezolid and tigecycline have demonstrated excellent in vitro activity against MRSA strains (Table 3). Only one HA-MRSA and multidrug resistant strain was classified glycopeptid-intermediate S. aureus (GISA) by MIC (vancomycin MIC = 1 µgml-1 and teicoplanin MIC = 4 µgml-1) and h-VISA by Macromethod Etest (vancomycin MIC = 12 µgml-1 and teicoplanin MIC = 24 µgml-1). This strain was isolated from blood of a 26-years-old patient with nosocomial bacteremia acquired in an intensive care unit who failed teicoplanin and vancomycin therapy. Only one CA-MRSA strain was resistant to tigecycline (MIC = 1 µgml-1). This strain caused skin and soft tissue infections (SSTI).
Table 3: MIC distributions and activities of vancomycin, teicoplanin, linezolid and tigecycline against the 135 MRSA strains. View Table 3
Among the 135 strains studied, 131 (48 CA-MRSA and 83 HA-MRSA) were typeable by smaI macrorestriction. Employing a cut off similarity value of 80% in subsequent cluster analysis, we assigned the HA-MRSA strains to 61 different PFGE patterns. HA-MRSA strains belonged to the same pattern were isolated in different hospitals. In addition, there is no dominant clone suggesting that these strains were not closely related (Figure 1). However, twenty different pulsotypes were identified among the 48 CA-MRSA strains. Three patterns identified in different hospitals were dominant: pattern 9 (n = 7), pattern 10 (n = 8) and pattern 11 (n = 8) (Figure 2).
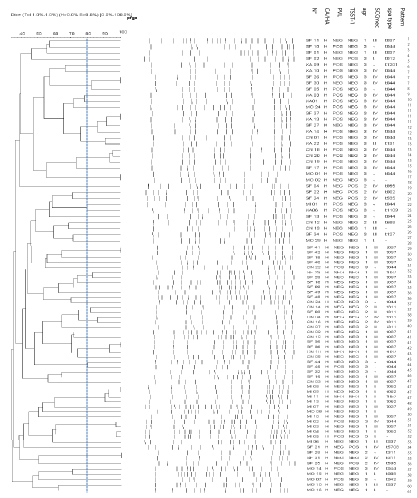 Figure 1: Dendrogram of PFGE patterns of HA-MRSA strains with genetic characteristics (PVL, TSST-1, agr, SCCmec and spa type) following digestion with sma I restriction enzyme.
Figure 1: Dendrogram of PFGE patterns of HA-MRSA strains with genetic characteristics (PVL, TSST-1, agr, SCCmec and spa type) following digestion with sma I restriction enzyme.
CA: Community-acquired; HA: Hospital-acquired.
View Figure 1
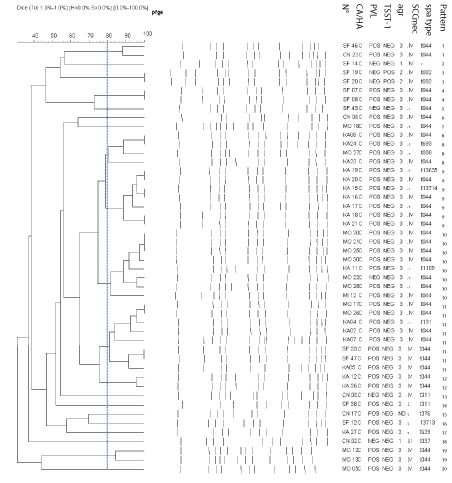 Figure 2: Dendrogram of PFGE patterns of CA-MRSA strains with genetic characteristics (PVL, TSST-1, agr, SCCmec and spa type) following digestion with sma I restriction enzyme.
Figure 2: Dendrogram of PFGE patterns of CA-MRSA strains with genetic characteristics (PVL, TSST-1, agr, SCCmec and spa type) following digestion with sma I restriction enzyme.
CA: Community-acquired; HA: Hospital-acquired.
View Figure 2
All MRSA strains carried mecA gene, which was detected by the PCR assay. The pvl gene was detected in 69 (51.1%) strains, 41 (83.7%) among CA-MRSA and 28 (32.6%) among HA-MRSA. Forty-five (69.6%) of pvl-positive MRSA strains were obtained from SSTI especially panaris (n = 17), phlegmon (n = 7), cutaneous abscesses (n = 5) and furuncle (n = 2).
The molecular characteristics (spa types, agr groups, SCCmec, presence of pvl gene, along with the drug resistance pattern of the most predominant types according to the origin of case (CA or HA) are summarized in Table 4.
Table 4: Molecular characteristics of MRSA strains isolated in Tunisia. View Table 4
Thirteen spa types were identified among CA-MRSA strains. The most common spa type was t044, 34 (69.4%). A combination of the molecular typing results revealed that thirty-two (78%) among pvl-positive CA-MRSA strains, belonged to spa type t044, have agr3, and most (n = 27) harbored SCCmec IV. These strains were often resistant to kanamycin, tetracycline and fusidic acid (Table 4). All of these characteristics were consistent with those of the European clone ST80. PFGE typing showed that among t044 strains, 41.7% were grouped into three dominant patterns (Figure 2).
Among HA-MRSA strains, 18 spa types were identified. The most prevalent were t037 (n = 25, 29.1%), followed by t311 (n = 7, 9.1%) and t052 (n = 5, 5.8%) among pvl-negative strains and t044 (n = 22, 25.6%) among pvl-positive strains. Further molecular characterization showed that all of the strains belonging to spa type t037 had agr1 and carried SCCmec III. In term of drug resistance, all of them exhibited resistance to kanamycin, tobramycin, gentamicin and tetracycline (Table 4). These characteristics were consistent with the ST239/247 clone.
Of the strains tested, only eight (5.9%) were tst-1 positive. They caused a variety of clinical syndromes and were associated to both CA infection (two endocarditis) and HA infection (three SSTI, two prosthesis infections and one bacteremia). These strains were not multidrug resistant: resistant to fusidic acid (n = 5), tetracycline (n = 1) and trimethoprim + sulfamethoxazole (n = 1) and were not closely related after analysis by PFGE. Four spa types were identified, t002 possessed agr2 and SCCmec IV (n = 3); t535, agr2, SCCmec IV (n = 2); t012, agr3, SCCmec I (n = 1) and t5708, agr1, SCCmec IV (n = 1).
MRSA strains continue to be isolated from both healthcare-and community-associated infections in different parts of the world [3]. The increase in the number of MRSA infections reported worldwide has been accompanied by changes in the characteristics of MRSA strains emerging in different parts of the world. Consequently, epidemiological typing using a combination of phenotypic and genotypic typing methods has contributed for understanding the evolution and dissemination of MRSA clones [15,23]. To date, few studies have been focused on the phenotypic and genotypic characteristics of MRSA strains in Tunisia [24,25]. This is the first multicenter study of molecular epidemiology focusing on CA and HA MRSA in Tunisian hospitals. The rate of MRSA in Tunisia was relatively low: about 20%. However, a slightly increase was showed (18.4 in 2004; 22.7% in 2014) [26,27].
In our study, 36.3% of MRSA strains caused CA-infection. The prevalence of CA-MRSA varies geographically; with detection rate remain low in most European countries: ranges from less than 1% in Switzerland to less than 10% in Austria and 7% in Russia [28,29]. This rate is highest in other countries such as Colombia (12.6%) [30], Kuwait (17%) [31], Algeria (34.3%) [32], Saudi Arabia (59%) [31] and Argentina (62.9%) [7]. CA-MRSA strains predominantly infect young and previously healthy patients. They cause mainly SSTI and some strains are particularly virulent and induce life-threatening infections such as necrotizing pneumonia or invasive osteomyelitis [2,3,7,31]. In our study, CA-MRSA strains were more frequently isolated from younger patients than HA-MRSA strains (p < 0.001). SSTI were significantly predominated in CA-MRSA infections (77.6% versus 36%; p < 0.001). However, bacteremia, respiratory tract infections and catheter related infections were significantly related with HA-infections.
Antimicrobial susceptibility testing is a crucial step in selection of the appropriate antibiotic for treatment. Surprisingly, the rates of resistance to tobramycin, gentamicin, rifampicin and trimethoprim-sulfamethoxazole among MRSA strains found in the present study were lower than that reported in previously studies in Tunisia [26,27]. This event could be explained by the emergence of CA-MRSA strains in our hospitals.
Our Study showed a low rate of resistance to antibiotics in CA-MRSA strains except for kanamycin (83.7%), tetracycline (75.5%), and fusidic acid (75.5%). Only 2% of the CA-MRSA strains were resistant to lincomycin. Then, lincosamides (clindamycin and lincomycin) remains a useful option for outpatient treatment. HA-MRSA is generally multidrug resistant [33]. In our study, more than 45% of HA-MRSA strains were resistant to aminosides, erythromycin, ofloxacin, tetracycline and fusidic acid (Table 2). Four strains (2.9%) were resistant to pristinamycin. In Tunisia, this resistance was rare [26,27] and first emerged among S. aureus in 2007 in Sfax university hospital [34].
In our study, only one HA-MRSA and multidrug resistant strain (0.8%) isolated from blood was classified as GISA by MIC, and h-VISA by macroEtest method. In Tunisia, GISA strains were uncommon [26,27,35]. Previous studies have demonstrated that h-VISA/GISA are prevalent among bacteremic specimens, and that these strains can persist in the blood stream for a long time [14]. In the literature, the incidence of GISA is low [13,14,36]. However, the rate of h-VISA among MRSA strains varied from countries (13% in Australia, 32% in Turkey, 11% in France and 7% in China) [14,37] and was parallels with the increase of vancomycin MIC level [38]. Vancomycin resistant S. aureus, due to the acquisition of the vanA gene from enterococci are currently very low [36,39,40]. Although the prevalence of GISA is very low in our country (0.8% among MRSA), a continuous surveillance of susceptibility to glycopeptides is necessary particularly among MRSA strains.
Systematic review and meta-analysis of genetic backgrounds of h-VISA/GISA demonstrated that the majority of these strains harbored SCCmec II or III, rarely SCCmec I and IV and belonged to ST239 and ST5 [14,40,41]. In our study, the h-VISA strain contained SCCmec I and was not typeable by spa typing.
Linezolid and tigecycline have demonstrated excellent in vitro activity against S. aureus including MRSA. Only one strain was resistant to tigecyclin (MIC = 1 µgml-1). Thus, they constitute a therapeutic alternative in severe infections especially due to GISA or h-VISA strains [33,42,43]. However, emergence of linezolid resistance in S. aureus has been reported in USA, in Europe and in Asia. This resistance was observed following exposure to linezolid [44-46].
A high rate of pvl-positive MRSA (50.4%) was found in our study especially in the community (83.7%). This finding has been reported in other studies [28,32,47-51]. pvl gene was strongly associated with SSTI especially panaris, phlegmon and cutaneous abscesses. In the literature, although the majority of MRSA-pvl positive infections are SSTI, sever life-threatening cases of necrotizing pneumonia and necrotizing fasciitis have been reported [1,3]. Fifty-four (78.3%) of the pvl-positive MRSA strains have characteristics of CA-MRSA European clone ST80-IV (spa type t044, agr3, SCCmec IV, pvl +). This clone accounted for 65.3% of CA-MRSA and 25.6% of HA-MRSA, thus proving its current emergence as an HA-pathogen.
CA-MRSA is particularly well established in the USA. Although USA400 (pvl-positive ST1 SCCmec IV) strains were responsible for initial cases, USA300 (pvl-positive ST8-IV) is now the predominant cause of North American infection and has begun to replace health-care-associated strains [5,23,28,31,52]. By contrast, USA300 is uncommon in Europe, where CA-MRSA is characterized by clonal diversity [48,50,53]. The most common European CA-MRSA clone is pvl positive ST80-IV, which have a characteristic antimicrobial susceptibility pattern of resistance to kanamycin, tetracycline and fusidic acid. European clone ST80 was reported in Greece since 2003, the country with the highest CA-MRSA incidence in Europe, where it is responsible for the majority of both CA-MRSA and HA-MRSA infections [53]. The European clone has been reported in many European countries [28,48,53], however, it is not very common in Slovenia [54]. CA-MRSA ST80 clone has been also identified in other countries from Africa, particularly in Algeria [32,55] and in the Middle East such as Kuwait, Lebanon, Jordan and Egypt [49,51].
Since their emergence, CA-MRSA ST80 strains were susceptible to several antibiotics and are usually resistant to kanamycin, tetracycline and fusidic acid [28]. However, new strains appeared to develop resistance to other antibiotics. In our study, among the CA-MRSA strains identified, we found multidrug resistant pvl-positive strains, showing resistance to other antibiotics such as rifampicin, gentamicin and ofloxacin as in the Algiers study [32]. These results prove that CA-MRSA are able to expand their resistance profiles in hospital setting where volumes of antimicrobial consumption are higher than in the community.
In recent years, several CA-MRSA clones have arisen in Europe, most notably the LA-MRSA, CC398, which was first detected in the Netherlands and Denmark in 2003 [8]. MRSA CC398 has been reported mostly in Europe and also in Asia and in the USA. It accounted for only a small proportion ranging from 0 to 25% of human MRSA isolates depending on regional differences in livestock density [5,8]. Methicillin-susceptible S. aureus belonging to CC398 have also been reported from animals and humans and have been associated with CA and HA infections in humans, many without livestock contacts [56-58]. Studies have revealed that LA-MRSA CC398 originated in humans as methicillin-susceptible S. aureus and then spread to livestock, where it acquired resistance to methicillin and tetracycline [57-58]. To data, in Tunisia, only one MRSA ST398-spa type t899 isolate in the nasal sample of a farmer patient was reported, representing the first report of ST398 in humans in North Africa in our knowledge [59].
Among HA-MRSA strains, twenty-five (29.1%) have the following characteristics: pvl-negative, spa type t037, SCCmec III and agr1. These strains were related to the Brazilian/Hungarian clone of HA-MRSA ST239 and ST like (ST239/240/241). This clone is an epidemic clone responsible for several HA-MRSA outbreaks. It has been found to be a cause of HA-infection in African countries including Algeria [55], Ghana, Morocco, South Africa, Nigeria and Kenya [23,60-62], and has been reported in the USA, Europe, Austria [63], Russia [29], Turkey [41], Asia, Argentina [7], China [64], and Romania [50]. This clone was also identified in the Middle East [65-68]. In fact, its broad distribution may be due to its advantageous properties with respect to other clones, such as an enhanced ability to form biofilm and a tendency to acquire genes that confer resistance to different classes of antimicrobial agents [29]. In this study, all of the HA-MRSA t037 strains were multidrug resistant, which is in agreement with previous reports [7,29,41,55,60-65].
Other spa types were also detected in our study such as t311 (n = 9) and t052 (n = 5) especially among HA-MRSA, pvl negative strains. t311 was also detected in CA and HA MRSA strains in previous study conducted in Military hospital of Tunis [59] and in some African towns in Morocco, Cameroun and Senegal [61]. ST5-II/t311 is a major prevalent clone that is widespread in a tertiary hospital in China (45.8%) [64], and in Argentina [4,7]. In our study, the five t052 strains were isolated from the military hospital of Tunis. All of them harbored SCCmec I and agr1 and showing the same profile (kanamycin, tobramycin, gentamicin, tetracycline, erythromycin, lincomycin, ofloxacin and rifampicin resistance). These strains were related to ST235/247 clone. This finding has also been reported in a previous study conducted in the military hospital of Tunis [59].
We also identified other minor spa types amongst CA-MRSA and HA-MRSA accounting for 2% each (one strain) such as t002, t008, t131, and t376 which have only one repetition of difference with spa type t044. These spa types were also described in many parts of World accounting for less than 10% among MRSA [4-7,30,52,64,69]. spa type t012, t127, t688 and t535 were rarely reported [6,33,64,66].
In our study, the tst-1 gene was detected in only 8 strains (5.9%). There have been few reports of MRSA isolates producing tst-1 in Japan, Germany and in France [70]. However, high prevalence of tst-1 gene among MRSA was found in Gaza hospitals (27.4%) [66] and in Kuwait hospitals (62.2%) [71]. tst-1 positive MRSA strains appear to be highly virulent and to cause a variety of illness, ranging from toxic shock syndrome to various suppurative infections [72] such as SSTI, pneumonia, osteoarthritis [70-73]. Few cases of endocarditis have been reported [74,75]. In our study, three patients had SSTI, two had prosthesis infection, two had endocarditis and one bacteremia. The tst-1 positive MRSA belonged to two clones: a major clone ST5 Geraldine clone, mainly spa type t002, and agr2, SCCmec IV, rarely SCCmec I, and one minor clone ST30, agr3, SCCmec IV [70]. However, in the middle East, other clone has been identified among tst-1 positive strains, ST22, SCCmec IV, t233 (middle Eastern variant) in Kuwait and Gaza hospitals [66,71]. In our study, only three strains have the characteristics of the Geraldine clone. One strain t5708, SCCmec, agr1 was also detected in Kuwait [71].
In conclusion, we reported that pvl-positive MRSA spa type t044 was spreading in both the community and hospital setting in different city of Tunisia. These strains have the characteristics of the European clone ST80 and were susceptible to most of the antistaphylococcal antibiotics and usually resistant to kanamycin, tetracycline and fusidic acid. However, the risk that multidrug resistant MRSA strains will spread to the community in Tunisia is a worrying prospect. spa type t037 clone which was multidrug resistant was found to be dominant in HA-infection. These findings support the need for measures designed to limit the spread of both CA and HA-MRSA, together with regular and systematic surveillance of MRSA infections in our country.
We thank Helene Meugnier (National Reference Center for Staphylococci, Lyon, France) for technical assistance.
There is no conflict of interest.