Strategies to improve time to administration of appropriate, effective antimicrobial therapy can improve patient outcomes. We sought to retrospectively assess if the earlier identification of blood pathogens and their resistance determinants with multiplex PCR platforms could have an impact on time to initiate appropriate antimicrobial therapy.
All patients with monomicrobial positive blood cultures from March to June 2013 were included in the retrospective chart review analysis. We assessed time to effective therapy (time from positive blood culture Gram stain result to change in therapy), time to optimal therapy (time from Gram stain result to the final change in therapy based on susceptibility), and compared to time to targeted therapy (time from rapid multiplex PCR results to modification of therapy).
One hundred and forty-nine patients were included. The average time to effective therapy was 7.6 hours, and time to optimal therapy 52.3 hours. Time to targeted therapy would be 1.15-2.5 hours with availability of multiplex PCR results (P < 0.001). A total of 28 patients would have received targeted therapy (1 with CTX-M K. pneumoniae, 1 with KPC K. pneumoniae, 3 with MRSA, 20 with MSSA and 3 with VRE infection) in significantly less time.
Use of the rapid multiplex PCR systems, had the greatest potential to improve timeliness to appropriate therapy in the patients where the presence or absence of drug resistance markers such as mecA, vanA/B, CTX-M, and KPC was determined.
Multiplex PCR, Blood cultures, Antimicrobial stewardship
Timeliness of antimicrobial administration has been shown to be associated with clinical outcomes such as length of stay and mortality [1]. Clinical guidelines for various infectious disease states including sepsis, febrile neutropenia, and candidemia emphasize the importance of timely administration of effective antimicrobial therapy, ideally within 1-2 hours of presentation [2-4]. While the timeliness of antimicrobial therapy can be influenced by many factors, laboratory diagnostics can play a major role in facilitating more expedient initiation of effective antimicrobial therapy [5].
For blood culture results, the traditional process of reporting organism identification and susceptibility results can be lengthy (2-4 days) and can delay optimal antimicrobial agent selection and initiation. Advances are needed to decrease the turn-around-time of organism identification and speciation from culture results. The solution is the availability of multiplex PCR platforms that can provide rapid identification and antimicrobial susceptibility results. The multiplex PCR blood culture platforms, FilmArray (FA) (BioFire, Salt Lake City, UT) and Verigene (NV) (Nanosphere, Northbrook, IL) offer accurate and rapid identification of multiple species of Gram-positive and Gram-negative organisms and can detect certain resistance markers such as mecA, vanA, KPC, CTX-M [6-16]. A previous study from our institution compared the ability of three systems and the time required to complete the identification: matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF), NV, and FA. It was observed that time to species identification was significantly reduced from an average of 25.6 hours (MALDI-TOF) to 1.15-2.5 hours with NV and FA. Moreover, the NV and FA systems accurately identified pathogens in 92% of cultures assessed [15].
While both the NV and FA offer similar benefit in regards to timeliness of results, there are important differences between the assays in terms of their targets. The NV Gram-positive blood culture assay can detect 9 bacterial species (S. aureus, S. lugdunensis, S. epidermidis, S. pneumoniae, S. anginosus, S. agalactiae, S. pyogenes, E. faecalis, E. faecium), 4 genera (Micrococcus spp., Staphylococcus spp., Streptococcus spp., Listeria spp.), and 3 genetic resistance determinants (mecA, vanA, and vanB). The NV Gram-negative blood culture assay can detect 5 different bacterial species (E. coli, K. pneumoniae, K. oxytoca, S. marcescens, P. aeruginosa), 4 genera (Acinetobacter spp., Enterobacter spp., Citrobacter spp., Proteus spp.), and 6 genetic resistance determinants (CTX-M, KPC, IMP, NDM, OXA, and VIM) [17]. The FA blood culture identification panel includes the same Gram-positive organisms and most of the same Gram-negative pathogens, plus Acinetobacter baumannii, Haemophilus influenzae, and Neisseria meningitidis and detects mecA, vanA, vanB, and KPC resistance determinants [18]. FA also provides identification of various Candida spp. (C. albicans, C. glabrata, C. krusei, C. parapsilosis, and C. tropicalis) [18].
In the present study we sought to retrospectively assess if the earlier identification of blood pathogens and genetic resistance markers with multiplex PCR platforms could have an impact on time to initiate targeted therapy (time from FA and NV results to when a modification of therapy would have theoretically occurred), compared to time to effective therapy (time to initiation of antibiotics based on Gram stain result). We separated the time frames to modify antimicrobial therapy into the three categories previously defined: targeted, effective, and optimized. We termed the time to respond to FA or NV results differently than the time periods assessed for the control group as the results of these platforms could enable for modification in therapy that would be congruent with the definition of effective therapy and/or optimized therapy, depending on what organism identification and resistance mechanism was identified.
This was a single-center retrospective evaluation, including all inpatients, regardless of age, with a positive mono-microbial blood culture between March and June 2013. This study was reviewed and approved by an institutional review board. Patient profiles were assessed to determine time to initiation of effective antimicrobial therapy and optimal antimicrobial therapy. Time to effective antibiotic therapy was defined as time from Gram stain report (date and time recorded with Gram stain indicating when the primary physician was contacted) to the time of administration of an antibiotic that had activity against the organism that ultimately grew in culture and to which the organism was susceptible to (e.g. if Gram-positive cocci on Gram stain and started on vancomycin). Time to optimal antibiotic therapy was defined as time from Gram stain report to the time the patient received antibiotic therapy (based on susceptibilities), which included de-escalation or escalation based on known susceptibility results, or dosing modification to improve efficacy (e.g. the organism had elevated minimum inhibitory concentrations (MICs). We also assessed theoretical time to targeted therapy based on if the NV and FA multiplex PCR platforms results had been available for the patients with positive blood cultures included in this study. Patients were excluded from the analysis if they had already been placed on effective therapy empirically, prior to the Gram stain result, or received targeted therapy prior to the availability of organism identification and susceptibility results, or not received antibiotics for the positive blood culture or expired prior to the availability of culture data. An evaluation of the two antimicrobial therapy-related outcomes (time to effective antibiotic therapy, time to optimal antibiotic therapy) was performed, comparing actual antibiotic administration times from chart review of patients with positive blood cultures in the absence of NV and FA multiplex PCR platforms to theoretical administration times based on the rapid identification of organisms and detection of resistance mechanisms provided by the multiplex PCR platforms, had they been available.
Microbiology reports were reviewed to identify patients for inclusion. Only mono-microbial blood cultures were included. All blood culture specimens were inoculated into Bactec culture bottles and incubated in the Bactec FX instrument (Becton Dickinsen, Cockeysville, MD), as per routine practice. A Gram stain was performed if a blood culture signaled positive for growth. Gram-positive, Gram-negative, and yeast pathogens were included. When cultures signaled positive, aliquots of broth medium were processed for detection by both NV and FA as well per manufacturing instructions. In brief: a sample from a positive blood culture bottle was injected into plastic pouch and placed in the FA multiplex PCR system, where sample preparation, amplification, detection and analysis are all integrated into one complete process. Similarly for the other assay, a sample from a positive blood culture, based on the results of the Gram stain, was injected into a either Gram positive or Gram negative cartridge, and placed into NV platform, where bacterial nucleic acid extraction, purification, microarray hybridization and signal amplification happen in a closed system. None of the results from the two platforms were communicated to providers or entered into medical record [15]. For the routine work up aliquots of the positive blood culture bottles were sub-cultured on agar plates, and following overnight incubation colonies were identified by Vitek MS RUO (BioMerieux, Marcy, L'Etoile, France). Susceptibility testing was performed as per routine protocol with Vitek 2 (BioMerieux, Marcy, L'Etoile, France). Time to identification was defined as time between Gram staining of the positive blood culture bottle and organism identification.
Statistical software (STATA, version 13.1) was used for statistical analysis. The Shapiro-Wilk test was applied to assess normalcy of data distribution. As the data was normally distributed, comparison of time to effective and targeted antibiotic therapy was performed using a paired sample t test.
One-hundred-and-fifty-six patients with mono-microbial blood cultures were initially evaluated; 7 were excluded that did not receive antibiotics for the positive blood culture result; of 149 included in the analysis, 93 had Gram-positive bacteria, 53 Gram-negative bacteria and 3 yeast (all Candida albicans).
Of the 149 patients assessed, 49 (33%) patients were assessed for time to effective therapy, 100 (67%) had already been started on effective antibiotics prior to the blood culture and Gram stain result and were excluded. Seventy-seven of the 149 included patients were assessed for time to optimal therapy; 72 (48%) were excluded as they had already been started on targeted therapy prior to availability of results). The observed average time to initiation of effective antibiotics was 7.6 hours and average time to optimal therapy was 52.3 hours. All 149 patients would have received targeted therapy in 1.15 hours if FA results were used or in 2 to 2.5 hours if NV results were used, assuming immediate response to results. For the purpose of this analysis we assumed immediate modification in therapy based on the FilmArray or Verigene results to a targeted antibiotic if antimicrobial stewardship intervention was utilized.
Shown in Table 1 is the breakdown of specific Gram-positive, Gram-negative, and yeast pathogens identified in culture among patients where further modification in antimicrobials was needed to implement optimal therapy. Of 53 patients that grew a Gram-negative pathogen, 51 had non ESBL/KPC isolates, with 28 requiring therapy modifications, after susceptibilities became available, 1 patient had ESBL producing (CTX-M) K. pneumoniae, and 1 KPC producing K. pneumoniae, both requiring therapy modification after susceptibilities were available. Of 93 patients that grew Gram-positive organisms, 39 grew Staphylococcus aureus in culture, 22 MSSA and 17 MRSA. Only 3 patients with MRSA and 20 with MSSA required therapy modification to more targeted therapy. Vancomycin resistant Enterococcus faecium was identified in culture for 5 patients; only 3 of these patients required therapy modification to targeted anti-VRE therapy. The respective average times to targeted, effective and optimal therapy and clinical outcomes assessed for each of these pathogens and groups of pathogens are shown in Table 1 and Figure 1.
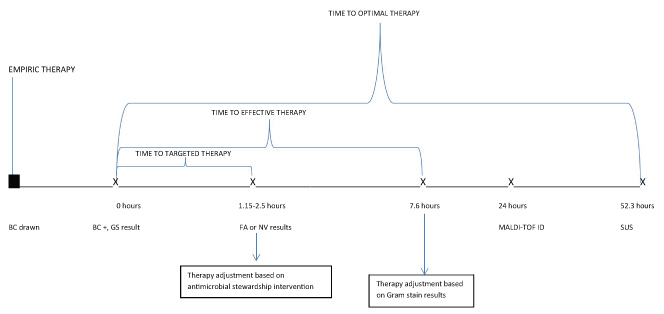 Figure 1: Result and Therapy Modification Timeline.
Figure 1: Result and Therapy Modification Timeline.
BC: Blood culture; GS: Gram stain; FA: Film Array; NV: Nanosphere Verigene; MALDI TOF: Matrix Assisted Laser Desorption/Ionization Time of Flight; ID: Identification; SUS: Susceptibility (full antimicrobial susceptibility results).
View Figure 1
Table 1: Time to Therapy. View Table 1
Based on previous data showing that FA and NV multiplex PCR platforms can identify Gram-positive and Gram-negative pathogens as well as select markers for resistance directly from the blood culture bottle within hours from the time of the Gram stain result, time to crucial modifications in antibiotic therapy can be drastically reduced compared to the traditional process of organism identification and susceptibility testing. For the cultures reviewed at our hospital, using the traditional process of MALDI-TOF identification, Vitek 2, and other phenotypic susceptibility testing, the overall average time to initiate effective antibiotic therapy based on Gram stain results was 7.6 hours, and time to tailored optimal therapy based on susceptibility results, was 52.3 hours. For those isolates identified specifically as MRSA, ESBL, or KPC-producers, the average time to targeted therapy would be approximately 1-2.5 hours, compared to the 52.3 hours it currently takes for optimal therapy using a traditional testing approach (P < 0.001).
With the rapid multiplex PCR systems, the ability to detect specific resistance markers such as mecA, vanA/B, CTX-M, and KPC had the greatest potential to improve timeliness to targeted therapy given that the presence of these genes quickly identifies a pathogen and the presence or absence of drug resistance. Twenty-two Staphylococcus aureus isolates would have been identified as being mecA-negative allowing de-escalation to antibiotic therapy targeted to MSSA (e.g. oxacillin or cefazolin) within 1.15 hours (FA) or 2 hours (NV) of the Gram stain result, versus at 52.3 hours. Five out of 11 Enterococcus species isolated in culture would have been identified as vanA positive within 1.15 or 2 hours of blood culture positivity, allowing for more timely modification in therapy to target VRE compared to the observed time of 48.7 hours. A patient with confirmed CTX-M positive K. pneumoniae (1 isolate) using the NV system (only this system can detect CTX-M) would have been able to be placed on targeted therapy within 2.5 hours, compared to the 29 hours for this specific case. Despite the relative delay to optimal antibiotic therapy, length of stay (LOS) was 3 days and the patient was alive at 30 days following culture result. For the patient with the KPC producing Klebsiella pneumoniae, the time to start optimal therapy was 33.9 hours, this patient's LOS was 68 days, and expired within 30 days of culture results. While it cannot be for certain whether quicker implementation of targeted therapy could have resulted in a better outcome, the availability of the KPC results within only a few hours of positivity of blood culture, would have significantly reduced time to optimal therapy which is known to be correlated with improved clinical outcomes [19,20].
The NV and FA systems may have a more limited impact on time to targeted therapy in cases where only species identification is available and when an effective antibiotic is already initiated based on Gram-stain results. In such cases, the identification provided the next day by MALDI-TOF did not result in a change in antibiotics in most cases. An example of this is identification of Candida albicans in blood cultures; despite our antibiogram showing favorable fluconazole activity against this specific pathogen, de-escalation to fluconazole from broader spectrum antifungal agents tends to not occur until susceptibility results are available. In the clinical setting, many physicians are reluctant to make modifications to more targeted therapy unless susceptibility results are available, especially in patients with clinical instability, immune dysfunction, and other comorbidities. Hence, it seems the greatest impact that our institution could expect from FA or NV is the rapid detection of specific resistance markers rather than just identification of the organism species alone.
Previous studies have evaluated the impact of the NV and FA systems on timeliness and appropriateness of antimicrobial therapy and on clinical outcomes. It is evident from the available literature that these systems can have a significant impact [19-26]. One study by Bork and colleagues evaluated the impact of the NV on the time to initiate effective (defined as time from Gram stain to when initial antibiotic started with activity against the isolated pathogen) and optimized therapy (time from Gram stain to when antibiotics were modified to more optimally cover the pathogen) for Gram-negative isolates. It was observed in this analysis that average time to effective therapy was reduced by 3.7 hours, and average time to optimal therapy was reduced by 18.3 hours [21]. Another study evaluated the impact of the NV Gram-positive blood culture PCR on antibiotic utilization for VRE and MSSA isolates, and found that average time to optimal antibiotics from the date of blood culture being drawn (initiation of oxacillin/nafcillin or cefazolin for MSSA, and daptomycin or linezolid for VRE) was reduced by 18.9 hours. Average time to optimal therapy for VRE isolates was reduced by 20.7 hours and MSSA isolates reduced by 20.6 hours [22]. Similarly Box, et al. found a significant reduction in average time to targeted therapy (time form empiric antibiotic to the time with known susceptibility results, with either escalation or de-escalation) by 25.7 hours [23]. Suzuki, et al. evaluated time to initiation of appropriate antibiotics from time of blood culture following the implementation of the NV Gram-positive and Gram-negative culture tests, and found a significant reduction as well [19]. For the FA platform, Ward and colleagues compared timing of NV results and FA results relative to traditional methods and determined that FA along with NV identified organisms 29.2 and 28 hours earlier than conventional methods, respectively [24]. Two studies have looked at the impact of time to antibiotic therapy for specific pathogens with the FA platform. For VRE blood culture isolates, MacVane, et al. observed a reduction in time to effective therapy (defined as time from blood culture draw and receipt of antibiotic with activity against patient's VRE isolate, and activity defined by susceptibility results), targeted to VRE bacteremia, was reduced from an average of 50.3 hours to 20.8 hours using FA multiplex PCR [20]. Another study looked at impact of FA on the time to administration of active therapy against MRSA, MSSA, VRE, and Candida spp. (time to active therapy was defined as time lapsed until susceptibility results were available. The results from this evaluation showed that patients with VRE bacteremia received active antibiotics 16 hours earlier, patients with MSSA received durations of unnecessary vancomycin that were 52 hours shorter, however time to effective therapy for MRSA and Candida spp. isolates was no different from standard culture and susceptibility methods with respect to time from collection or from time of positivity to time of therapy modification [25]. Bannerjee, et al. evaluated the impact of multiplex PCR based blood culture identification and susceptibility testing alone or combined with antimicrobial stewardship review on antimicrobial usage and de-escalation. Compared to the control arm the use of the FilmArray Blood Culture ID panel resulted in a reduction in the use of broad spectrum antibiotics. They also observed lower rates of treatment of contaminants, and time from gram-stain to de-escalation or escalation was reduced. The addition of antimicrobial stewardship review of the PCR results further reduced time to de-escalation/escalation (21 hrs/5 hrs vs. 38 hrs/6 hrs) [26. Our data showed that very few patients with MRSA needed further modification in therapy, therefore would expect that these multiplex PCR platforms would have little impact on time to optimal therapy for MRSA as this study found. Additionally, while we had only 3 Candida spp. isolates and all were C. albicans, none of the patients had their therapy modified until susceptibilities were reported, thus these rapid multiplex PCR platforms would also not be expected to impact time to targeted therapy for candidemia.
One of the limitations of this analysis is the retrospective nature of the study. Having to review charts to assess reasoning behind changes in antibiotics is often limited by detail and accuracy of information provided in medical charts. However, it was evident based on timing of antibiotic changes relative to the timing of Gram-stain result, culture identification, and susceptibility results whether the change was a result of the availability of the information provided by the data. Another limitation is that we also assumed the impact of the FA and NV results resulted in changes immediately upon notification of the result. While certainly our data shows that effective therapy was not initiated until 7.6 hours of Gram stain result suggest that with actual implementation clinically, a delay is expected; however, even if additional delay occurred after the multiplex PCR result, the time to targeted therapy would still be significantly improved from the average of 52.3 hours that we observed using the traditional method. It seems that the greatest impact would have been achieved if results were delivered directly to the antimicrobial stewardship team that could act upon them immediately. Additionally, clinicians tend to act more quickly when escalating and initiating alternative targeted antibiotics (i.e. changing therapy from cefepime when CTX-M is detected) rather than when de-escalating (except in cases of MSSA, where clinicians tend to de-escalate therapy quickly). We also are assuming that all cases in which therapy needed to be modified that the susceptibility results would not then identify need to modify therapy further subsequent to FA and NV results. In the case where for example therapy is modified based on the detection of the vanA gene with an Enterococcus faecium isolate, so vancomycin is transitioned to daptomycin. But if susceptibility testing revealed daptomycin resistance, then the time to targeted therapy for this organism would be no different than the traditional process as further modification to targeted therapy for daptomycin non-susceptible VRE would have been needed following availability of susceptibility data. Multiplex PCR platforms which provide rapid identification of species and presence of mecA, CTX-M, KPC, and other determinants for gram-negative resistance can significantly reduce time to targeted therapy. Based on our assessment, modifications in therapy predominantly occurred when susceptibility results were available. We believe that the greatest impact of multiplex platforms will be the ability to quickly identify genetic determinants for resistance (e.g. mecA negative Staphylococcus aureus or CTX-M positive Escherichia coli). These results using the FA or NV would enable escalation to targeted therapy within 1-2 hours from Gram stain result depending on the platform used, compared to the > 50 hours to optimal therapy as observed in our study using the traditional process in a setting were modification in therapy to a specific microorganism usually does not occur until susceptibility data is available. It is unclear based on our observed clinical outcomes in comparison to that of previous studies, if mortality or LOS would be significantly improved with the availability of these multiplex PCR platforms.