Plasmid rep genes, Acinetobacter plasmids, Healthcare-associated infections, Opportunistic pathogen, Antimicrobial resistance genes
Acinetobacter baumannii is an important opportunistic pathogen responsible for a variety of nosocomial infections [1,2]. Its success in the hospital environment obeys to multiple causes, among them, the ability to resist antimicrobial compounds. It can rapidly evolve Multidrug Resistance (MDR) when confronted with antibiotic therapy [1-3] and in particular, the emerging resistance to last-resort carbapenems represents a major concern worldwide [3]. The most frequent cause of carbapenem resistance in A. baumannii is represented nowadays by the acquired Carbapenem-Hydrolyzing Class D β-Lactamases (CHDL) of the OXA-23, OXA-40 and OXA-58 groups, with the respective blaOXA genes generally embedded in distinct genetic structures carried by plasmids [2,4-7].
Plasmids are extrachromosomal elements playing key roles in the dissemination of antimicrobial resistance genes and other adaptive determinants among resident pathogens in the clinical setting [8,9]. They are generally built up by the juxtaposition of functionally different genetic modules of various phylogenetic origins. A considerable part of the plasmid structure corresponds to essential modules related to replication and mobilization (the plasmid backbone), separated from others conferring adaptive functions to the host (the adaptive module) [8-10]. The former modules determine the range of hosts in which the plasmid could survive, and in this context many plasmids (Acinetobacter plasmids among them) show preferences for phylogenetically-related bacterial hosts [9-12]. Since plasmid backbone synteny is much more conserved than that of adaptive modules which rapidly vary depending on selective pressures, typing schemes based on replication and/or mobilization modules are amply used for plasmid classification [9]. An informative plasmids-classification system for a given bacterial population certainly contributes to the understanding of their dynamics and preferred routes of propagation of adaptive modules carrying antimicrobial resistance genes, an information allowing measures to control their dissemination [9].
Replicons are the minimal plasmid portion that replicates with the typical copy number of the parent plasmid. They are constituted by an origin of replication (ori) and genes encoding replication initiator proteins (Rep) that bind to the ori site [9,13,14]. A series of tandem repeat sequences known as iterons are part of the ori, and play a crucial role in plasmid replication initiation and copy number control [9,15]. Sequence analysis of plasmid replicons corresponding to A. baumannii clinical strains revealed many differences with those from other bacterial species, strongly suggesting that A. baumannii contains its own plasmid types [9-12]. This lead to a classification scheme based on replicase (rep) genes comparisons to categorize A. baumannii plasmids into homology groups [11,12]. Thus, based on the comparisons of the nucleotide and corresponding translated amino acid sequences of 27 rep genes present in 23 A. baumannii plasmids, Bertini, et al. [11] originally defined 19, different homology Groups (GR). The use of this typing scheme indicated that blaOXA-23 genes were apparently restricted to plasmids assigned to a limited number of GR, whereas others such as blaOXA-40 and blaOXA-58 were more widespread with the latter associated to up to 9 different GR [7,11,12]. Since these works more A. baumannii plasmid sequences have been reported, and a recent phylogenetic analysis [16] based on the alignments of 50 A. baumannii replicase proteins (Rep) identified a new homology group defined as GR20 which is notably restricted to Acinetobacter "small" plasmids (i.e., less than 10 kbp).
We have recently sequenced and annotated the plasmids carried by A. baumannii Ab242, a MDR clinical strain displaying carbapenem resistance obtained from a public hospital of Rosario, Argentina [6,17,18]. MLST analysis assigned this strain to the sequence type 104 (ST104, Oxford scheme) of the Clonal Complex 104 (CC104), one of the CCs responsible for the dissemination of blaOXA-58 genes in South America [19,20]. Sequence analysis revealed the presence of three novel plasmids in Ab242: pAb242_9, pAb242_12 and pAb242_25 (GenBank accession numbers KY984045, KY984046 and KY984047, respectively). While both pAb242_9 and pAb242_12 lack antimicrobial resistance genes, pAb242_25 carries a 9,641-bp adaptability module harboring an ISAba825-blaOXA-58 arrangement responsible of the carbapenem resistant phenotype and also a TnaphA6 transposon conferring aminoglycoside resistance [6,21]. Sequence analysis and database comparisons of the four replicases encoded in Ab242 plasmids indicated that all of them belonged to the Rep-3 superfamily. However, a more detailed characterization of these proteins following the above-mentioned classification schemes [11,12,16], revealed that three of them could not be confidentially assigned to any of the presently-defined Rep GR. Thus, to further characterize the Rep proteins encoded in Ab242 plasmids, in this work we re-evaluated the current Rep-based strategy using replicases obtained from 215 A. baumannii plasmid sequences deposited in public databases.
A search for A. baumannii plasmid sequences in the GenBank database (February 2017) was performed with no imposed size limitation. A total of 215 nucleotide sequences, which included both complete and partial plasmids, were retrieved. A local database containing all predicted protein sequences encoded in these sequences was then constructed. Next, a BlastP search for Rep proteins among this protein database (using 85% sequence identity and 85% query coverage cut-off values) was performed (only best hits were considered) using as query one member of each of the 20 previously defined GR groups [11,16] plus those proteins annotated as replicases in Ab242 plasmids.
A multiple alignment of the amino acid sequences of 122 inferred Rep proteins (see above) was carried out using MUSCLE [22], implemented within the Molecular Evolutionary Genetics Analysis tool, MEGA version 7.0 [23]. This tool was also used to infer Rep proteins phylogeny using the Neighbor-Joining method. Evolutionary distances were computed using the Poisson correction method [24]. The reliability of the inferred tree was tested by the bootstrap technique with 100 replicates.
Tandem repeat sequences (iterons) were searched at each side (650 bp) of the identified Ab242 replicase genes using the Tandem repeat finder (TRF version 4.09, Boston University) on-line tool (https://tandem.bu.edu/trf/trf.html). Alignment of iterons sequences corresponding to each Rep group was then performed for comparative analyses.
Ab242 plasmids annotation inferred the existence of four rep genes, one each in pAb242_9 and pAb242_12 and two in pAb242_25. Encoded proteins belong to the Rep-3 superfamily similarly to most A. baumannii plasmid replicases [11,12,16]. Each of these replicases was individually used as a query in a BlastP-homology search against representative Rep proteins of each of the 20 presently defined GR [11,16]. This analysis could confidentially assign the only Rep encoded in pAb242_9 (locus tag pAb242_9_15) to GR4, as judged by the 95% sequence identity with RepAci4 from A. baumannii p844 (Table S1). On the contrary, the other three inferred Rep proteins (one in pAb242_12 and two in pAb242_25) showed non-significant identity and query coverage values (cut-off was set to 85% sequence identity and 85% query coverage) to any of the representative Rep proteins of the currently-defined GR [11], therefore precluding the assignment of these plasmids to any of these groups.
In an attempt to better characterize the aforementioned Ab242 plasmids on the basis of the Rep GR scheme, a local database containing all inferred protein sequences encoded by 215 plasmid sequences of A. baumannii deposited in databases was first constructed. Sequences ranged from 2.3 kbp (pTS236, Acc. no. JN872565) to 148.9 kbp (pAB3, Acc. no CP012005), and included both small cryptic to large self-transmissible plasmids [16,25]. Then, these plasmids were categorized on the basis of replicon typing [11] but employing a more restrictive BlastP search using as query representative Rep proteins of each of the presently-defined GR homology groups plus the three unclassified replicases encoded in Ab242 plasmids (see the Material and Methods section). This analysis identified 122 Rep-encoding sequences among the 215 plasmid sequences mentioned above (Table S1) indicating that a large proportion (58%) of the sequences analyzed lack distinguishable rep genes. This relatively large proportion of A. baumannii plasmid sequences lacking noticeable replicases could have resulted from the analysis of only partial plasmid sequences, in addition to the use of a restrictive cut-off value in our BlastP-based search (see above). It is worth noting, however, that some widespread A. baumannii plasmids might use a different replication strategy [16]. A phylogenetic analysis based on the alignments of the amino acid sequences of the above 122 Rep proteins was then conducted (Figure 1). The results of this analysis showed an overall clustering coincident with the A. baumannii plasmid Rep homology groups scheme proposed by Bertini, et al. [11], except for the differences observed at the level of GR8 (see below). In addition, our results (Figure 1) agree with the recent re-evaluation of this classification carried out by Lean & Yeo [16] who assigned a new homology Group (GR20) to this scheme. Interestingly, three of the four replicases encoded in Ab242 plasmids, one in pAb242_12 and two in the pAb242_25, did not fall into the previously defined groups [11,16] thus indicating the existence of novel GR (Table S2). Following the nomenclature used by these authors, we thus denominated the only replicase encoded in pAb242_12 as RepAci21 (locus tag pAb242_12_1), thus defining a novel GR21 group in the A. baumannii Rep-based plasmids classification scheme (Figure 1 and Table S2). In turn, for the two-replicon plasmid pAb242_25 one replicase was designated as RepAci22 (locus tag pAb242_25_21) and the other as RepAci23 (locus tag pAb242_25_24) thus defining the novel plasmid groups GR22 and GR23, respectively. Each of these two groups also include a second Rep member whose sequences were retrieved from databases, the replicases encoded by pD36-4 (Acc. no. ALJ89842) and p11921 (Acc. no. ADM89095), respectively. Although the latter protein had been previously assigned to Rep GR8 with a marginal confidence value [11], our analysis here now assigned it to the newly-defined GR23 with a branch bootstrap support of 100% (Figure 1).
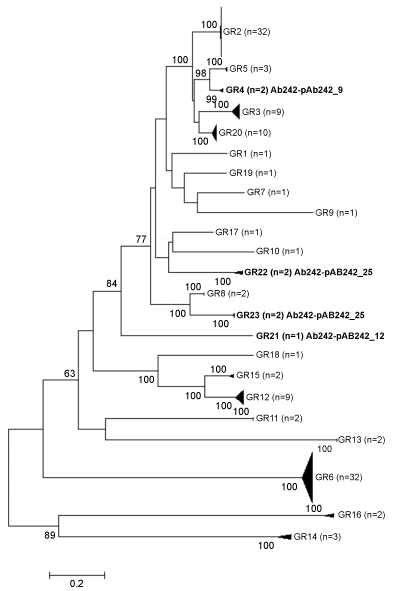 Figure 1: Unrooted NJ-phylogenetic tree constituted by 122 A. baumannii replicase proteins (see Table S1 and Material and Methods for details). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances (number of amino acid substitutions per site) were computed using the Poisson correction method [24]. The percentages of 100 bootstrap resamplings supporting the different clusters, as obtained by NJ is indicated at the bifurcations (bootstrap support values higher than 60% are indicated). For simplicity, the clades corresponding to each homology Group (GR) were collapsed and the number of members in each of them (n) is indicated. View Figure 1
Figure 1: Unrooted NJ-phylogenetic tree constituted by 122 A. baumannii replicase proteins (see Table S1 and Material and Methods for details). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances (number of amino acid substitutions per site) were computed using the Poisson correction method [24]. The percentages of 100 bootstrap resamplings supporting the different clusters, as obtained by NJ is indicated at the bifurcations (bootstrap support values higher than 60% are indicated). For simplicity, the clades corresponding to each homology Group (GR) were collapsed and the number of members in each of them (n) is indicated. View Figure 1
Iterons are tandemly repeated sequences located within the origin of replication initiation and part of the plasmid replication module [9,15]. The binding of a Rep protein to A. baumannii plasmids iteron sequences was demonstrated using pMAC from A. baumannii ATCC19606, reinforcing the notion that iterons play roles in the replication of these plasmids [26]. However, iteron number and spacing can significantly differ among plasmids [15], an observation that prompted us to search for putative iterons in the regions bordering each of the Rep-encoding genes in Ab242 plasmids (see above). This analysis detected the presence of 4 to 5 putative iterons each of 22 bp in length, located 56 to 220 bp upstream of the corresponding rep gene start codons (Table 1). The number and sequences of the direct repeats accompanying the RepAci4-encoding gene present in pAb242_9 were identical to those of pAb844 [11], both plasmids assigned to GR4 (Table 1). In the case of RepAci22 encoded in pAb242_25 and belonging to GR22, 4 repetitions were present next to the corresponding gene (Table 1). Notably, 3 repetitive sequences found in pAb242_25 sharing only 17 out of 22 nt were identified near the Rep-encoding gene present in pD36-4 [27] also part of GR22 (Table 1). Similarly, in the proximity of the RepAci23-encoding gene of pAb242_25, five highly similar repetitive sequences were present whereas 4 were observed for the other GR23 member, p11921 (Table 1). In line with these observations, Lean and Yeo [16] reported the presence of 3-6 repetitive sequences (19-22 bp in length) located 10-200 bp upstream of 50 rep genes analyzed. Altogether, these results suggest that each iteron sequence is characteristic of its cognate replicase gene.
Table 1: Replicon-associated iterons identified in GR members under study. View Table 1
Plasmids are important vehicles for the dissemination of resistance genes among nosocomial pathogens including A. baumannii [7-12]. In this context, plasmid typing is a useful tool for studying plasmid spread and evolution within a species population [8,9]. We updated here the A. baumannii plasmid classification scheme based on the clustering of plasmid-encoded replicase proteins [11,12,16]. By employing the comparisons of a larger set of 122 Rep amino acid sequences deduced from the analysis of 215 A. baumannii plasmid sequences, we revealed the existence of three novel Rep homology groups thus totalizing at present 23 different GR. Remarkably, the three Rep proteins that define these novel GR were encoded in plasmids detected in A. baumannii Ab242, a carbapenem-resistant clinical strain assigned to the CC104 (Oxford scheme) bearing a blaOXA-58 CHDL gene. The linking of blaOXA-58 to GR22 and GR23 reported here further supports the notion that this CHDL gene is more widespread among A. baumannii plasmids as compared to other blaOXA genes [7,11,12]. Also, and in agreement with other authors [16], we observed in our analysis a correlation between iteron sequences and a particular GR thus opening the possibility of using these repetitive sequences to refine the current Rep-based A. baumannii plasmid classification scheme.
This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT PICT-2011-1020) and CONICET (PIP 1055) to AMV; ANPCyT PICT-2012-0680 to ASL; and Ministerio de Ciencia, Tecnología e Innovación Productiva, Provincia de Santa Fe, Argentina, to A.M.V and A.S.L. M.M.C. is Fellow of CONICET, J.M.B., G.D.R and A.M.V. are Career researchers of CONICET, and A.S.L. is a Researcher of the UNR.