Exploration of single nucleotide polymorphisms (SNPs) that alter the expression or function of a gene may enable the development of diagnostics for endometrial cancer susceptibility.
We evaluated eleven candidate SNPs that have previously been reported in the literature or that are associated with cadmium sequestering (i.e., metallothionein) for their effects on endometrial cancer risk. We also predicted haplotypes for SNPs within genes on chromosomes 6, 14 and X and tested haplotype effects for cancer risk.
Our population-based study comprised 480 cases and 513 controls with 96% non-Hispanic White. Nine of the 11 SNPs were successfully assayed from the forward and reverse DNA strands. Concordance rate among genotype pairs was 99.73%. Our study found suggestive evidence of an increased endometrial cancer risk for two SNPs within the ESR1 and ESR2 genes (ESR1:rs2234693; ESR2:rs944050) but not for several other SNPs within those genes (ESR1:rs9340799; rs3020314 or ESR2:rs1255998). Likewise, an increased risk for carriers of two rare haplotypes (ESR1 haplotype:rs2234693:rs9340799:rs3020314) and (ESR2 haplotype:rs1255998:rs944050) was found using conditional logistic regression multivariable models. We found no association between SNPs located in metallothionein (rs2836603, rs3918249), progesterone receptor (rs1042638), androgen receptor (rs12011793, rs12011518), aromatase enzyme (rs4775936), or chemokine c-x-c motif (rs352038) and endometrial cancer risk.
Our data suggest that the ESR1 (rs2234693) and the ESR2 (rs944050) polymorphisms are associated with an increased risk of developing endometrial cancer. Further elucidation of genetic influence on endometrial cancer risk adds evidence to can inform precision medicine approaches.
Endometrial cancer, Cadmium, Single nucleotide polymorphisms, Estrogen receptors, Metallothionein
Endometrial cancer is the fourth most common cancer of women in the United States and occurs primarily in postmenopausal women. The American Cancer Society (2018) [1] reported that 63.230 women were expected to be diagnosed annually with cancer of the body of the uterus (endometrial cancers and uterine sarcomas), with 11.350 deaths annually. The 5-year relative survival rate is about 84% for white women and 62% for black women post-diagnosis (2018). Endometrial cancer is divided into two types. Type 1 tumors comprise the majority of diagnoses, with most being classified as adenocarcinomas. These tumors are associated with endogenous and exogenous estrogen exposure [2]. Type II tumors are primarily serous carcinomas and are often characterized as estrogen-independent. Type II tumors are believed to arise from atrophic endometrium and account for a disproportional number of deaths [2,3]. Cadmium, a toxic non-essential metal, mimics estrogen and may increase estrogen-receptor-mediated proliferation for the Type I cancer [4]. Other established risk factors for Type I cancer include obesity, nulliparity, and diabetes. Risk factors for Type II endometrial cancer are less well established [4,5].
According to Meyer, et al., [5] candidate single nucleotide polymorphisms (SNPs) may be involved in mechanisms that affect Type I and/or Type II endometrial cancer such as DNA damage repair, apoptosis, steroid receptor genetic pathways, carcinogen metabolism, steroid metabolism, and cell-cycle control. SNPs and/or haplotypes constructed from SNPs within candidate genes have been reported in a limited number of studies as being associated with endometrial cancer risk, though the findings are inconsistent [5,6].
In this study, we assessed the effects of genetic variants that had previously been identified as being significantly associated with endometrial cancer risk as well as SNPs associated with genes responsible for cadmium sequestration on risk of endometrial cancer. Of the 12 SNPs originally selected as candidates for endometrial cancer risk, rs1042638 (a 3' UTR variant in TPD52 with progesterone receptor gene PGR) was monomorphic and removed from further analysis. The remaining 11 SNPs were located within 7 genes: chromosome X: rs12011793 and rs12011518 (AR: androgen receptor); chromosome 6: rs2234693, rs3020314, rs9340799 (ESR1: estrogen receptor 1); chromosome 14: rs944050, rs1255998 (ESR2: estrogen receptor 2); chromosome 4: rs352038 (CXCL3: chemokine c-x-c motif, ligand 3); chromosome 20: rs3918249 (MMP9: matrix metallopeptidase 9); chromosome 15: rs4775936 (CYP19A1: cytochrome P450); and chromosome 16: rs28366003 (MT2A: metallothionein 2A). Individual SNP effects were investigated using single-marker analyses, and combined SNP effects were assessed by testing additive effects of haplotypes construction for the SNPs located within ESR1, ESR2 and ARon chromosomes 6, 14 and X, respectively.
Genetic data were collected from participants in a population-based endometrial case-control study conducted during 2011-2012. The design and participation proportions were described in detail elsewhere [7]. Briefly, ascertainment of incident endometrial cancer cases came from three cancer registries: Arkansas Central Cancer Registry, Iowa Cancer Registry and Missouri Cancer Registry and Research Center. Each registry contacted potential cases to gain written permission to pass their names to Health and Environmental Exposure (HEER) study staff. Using available data from Iowa and Missouri voter registration lists, controls were randomly selected within age strata. For each case, four controls with the same birth date (Iowa) or approximate age (Missouri) were sent invitation letters. Ineligible participants included those who contacted HEER staff to decline participation, those in which a telephone number or current address could not be found.
Trained interviewers called potential participants to obtain information on known and suspected risk factors. The age at, or year of, cancer diagnosis for cases was used as the reference age, or year, for the age-matched controls, respectively. At the conclusion of the interview, mailing addresses were confirmed and saliva and urine collection kits were mailed to the interviewed participants for home collection. These samples were returned to the study center for processing in pre-stamped addressed envelopes that were included with the collection kits. Participants who returned urine kits were sent a $25 check as an indication of appreciation. This study was approved by the University of Missouri Health Sciences Institutional Review Board as well as the review boards of the respective state cancer registries. Each participant provided informed consent.
Urine cadmium content was quantified by using inductively coupled plasma-dynamic reaction cell-mass spectrometry (ICP-DRC-MS) following the established protocol from the National Health and Nutritional Examination Study (NHANES) [8] with an additional correction for the effect of strontium on the tin interference. A comprehensive quality-control program incorporating numerous methods including bottle blanks, monitoring multiple cadmium isotopes, internal and external controls, stability samples, replicates, spikes, and routine inclusion of National Institute of Standards and Technology (NIST) standard reference materials was utilized to ensure high-quality data. The method detection limit of 0.0082 μg/L cadmium enabled quantification of metal levels in every urine sample. Urine creatinine level was measured using a colorimetric assay based on the Jaffé reaction to control for kidney function [9] See McElroy, et al. [7] for more details about cadmium quantification.
Participants collected saliva samples using the Oragene DNA/saliva kit disk format (DNA Genotek, Ottawa, Ontario) and mailed the sample to the study center. For processing, the vials were placed in an air incubator at 50℃ overnight. Next, they were mixed with 500 μl of Oragene* DNA Purifier and incubated on ice for 10 minutes. After incubation, they were centrifuged at 15.000 × g for 5 minutes at room temperature. To precipitate the DNA, the supernatants were recovered, mixed with 500 μl 100% ethanol and centrifuged at 15.000 × g for 5 minutes at room temperature. The recovered pellets were then washed with 250 μl 70% ethanol and centrifuged at 15.000 × g for 5 minutes at room temperature. The final step included rehydrating the DNA pellets by adding 100 μl of TE buffer (10 mM Tris-HCL, 1 mM EDTA, pH 8.0) and incubating the pellets at 50℃ for 1 hour with occasional vortexing.
DNA concentration and quality was assessed using a NanoDrop™ 2000 spectrophotometer (Thermo Scientific, Waltham, MA). Samples identified as having an A260/280 optical density ratio of less than 1.8 were re-extracted using a standard phenol-chloroform isoamyl alcohol extraction. Aliquots of 500 ng of DNA were sent to GeneSeek (a Neogen Company, Lincoln, NE) for SNP genotyping.
Genotypes for the 11 variable SNPs located within 7 genes (1 CXCL3, 3 ESR1, 2 ESR2, 1 CYP19A1, 1 MT2A, 1 MMP9, 2 AR) were obtained using a Sequenom Mass ARRAY® Analyzer 4 at GeneSeek. Each SNP was assayed twice with detection probes designed from each DNA strand. For SNPs located within the same gene, haplotypes were phased and 146 missing values (2.1% of the data) were estimated using Beagle 4.0 (Browning and Browning, 2007) to produce complete data for the loci (ESR1hap rs2234693:rs9340799:rs3020314, ESR2hap rs1255998:rs944050 and ARhaprs12011793:rs12011518). Allele frequencies were estimated by allele counting. Genotypes were tested for departures from Hardy-Weinberg equilibrium (HWE) using a chi-square test with Bonferroni correction for multiple testing.
We determined associations between SNPs and case-control status after controlling for other known risk factors for endometrial cancer. To determine which participant characteristics were associated with endometrial cancer, we used chi-square and t-test analyses to examine the association of categorical and continuous survey variables, respectively, with case-control status. We analyzed additional survey variables in categories (e.g., medical history, demographics, dietary habits) to determine which were the most strongly associated with case-control status. Variables with P < 0.1 that had the strongest association within categories were then selected as additional candidates for inclusion in the multivariable conditional logistic regression analysis. Model variables were selected by both forward stepwise inclusion and backwards elimination to determine if the same factors were chosen, using P < 0.1 for both model entry and exit criteria. Using all available survey data, we developed a multivariable conditional logistic model for case-control status, stratifying on age at diagnosis. Interactions between variables in the model were tested; none were significant and were thus not retained.
Stepwise and backwards variable selection retained the same set of variables, yielding a final set of independent variables of: Age at diagnosis; non-Hispanic African-American race; marital status; body mass index (BMI); history of trying to lose weight; smoking status; pack-years of smoking; history of endometriosis, breast cancer, ovarian cancer, or uterine fibroids; endometrial cancer in a first degree relative; years of oral contraceptive use; years of unopposed estrogen use; menopause status; menopause after age 56; number of protein shakes consumed per week; and 5 or more servings per week of whole milk.
We used Fisher's exact test to compare genotype frequencies between the cases and controls. With log-transformed creatinine-adjusted cadmium concentration as the dependent variable, we tested the association of 11 SNPs and 3 loci with cadmium in simple regression models. Each SNP and locus was subsequently included as a covariate in a conditional logistic model for case-control status that included the selected covariates described above as well as log-transformed cadmium concentration. For those loci for which a statistically significant association with urine cadmium levels was found, cadmium was excluded from the multivariable model used in the case-control analysis.
Because not all women returned their home biospecimen kits, and because women who did and did not return these samples differed on several characteristics (for example, alcohol consumption, medical history, education, and income), we developed a multivariable logistic model for the probability of an eligible participant returning a kit. The probability of returning a sample predicted by this model was used to adjust for non-response bias in the final case-control models by including the inverse of the probability as a weight [10].
Genotype data were added to the conditional logistic regression models to determine if SNPs or the loci were associated with case-control status after controlling for other risk factors. We tested the additive effects of SNPs by fitting a model with genotypes treated as allele dosages (e.g., CC = 0, CT = 1, TT = 2) with increasing dosages representing increasing numbers of copies of the rare (minor) allele at each locus. We tested non-additive effects at each SNP using two variables (a and d) following the notation of Falconer (1960) [11]. Samples with 0 copies of the common allele were coded as a = -1, d = 0; those with 1 copy were coded as a = 0, d = 1; and those with 2 copies were coded as a = 1, d = 0. Three SNPs (rs352038, rs944050, and rs12011518) had very rare minor alleles that prevented convergence of the logistic models, and were excluded from the non-additive effect analyses. Additive analyses were also performed for the haplotype effects by forming dummy variables for each haplotype in which the number of copies (0, 1, or 2) of each haplotype present in each diplotype (genotype based on haplotypes) were scored. For some diplotypes and haplotypes that were rare, these were dropped from the analysis.
While missing phenotypic data were rare, data imputation is considered to result in less bias than only analyzing cases with no missing data [12]. We therefore imputed missing phenotypic values using multiple imputation in SAS to create 20 datasets using the Markov chain Monte Carlo method. All final multivariable models for case-control status were run using the 20 imputed datasets and the MIANALYZE procedure in SAS was used to summarize results. Data were analyzed using SAS for Windows 9.4 (Cary, NC).
Sample derivation has previously been described [7] and is summarized in Figure 1. Briefly, of the 1519 women who completed the telephone survey, 517 failed to return analyzable saliva samples. We excluded nine controls that reported a history of endometrial cancer, leaving 480 cases and 513 controls in the study.
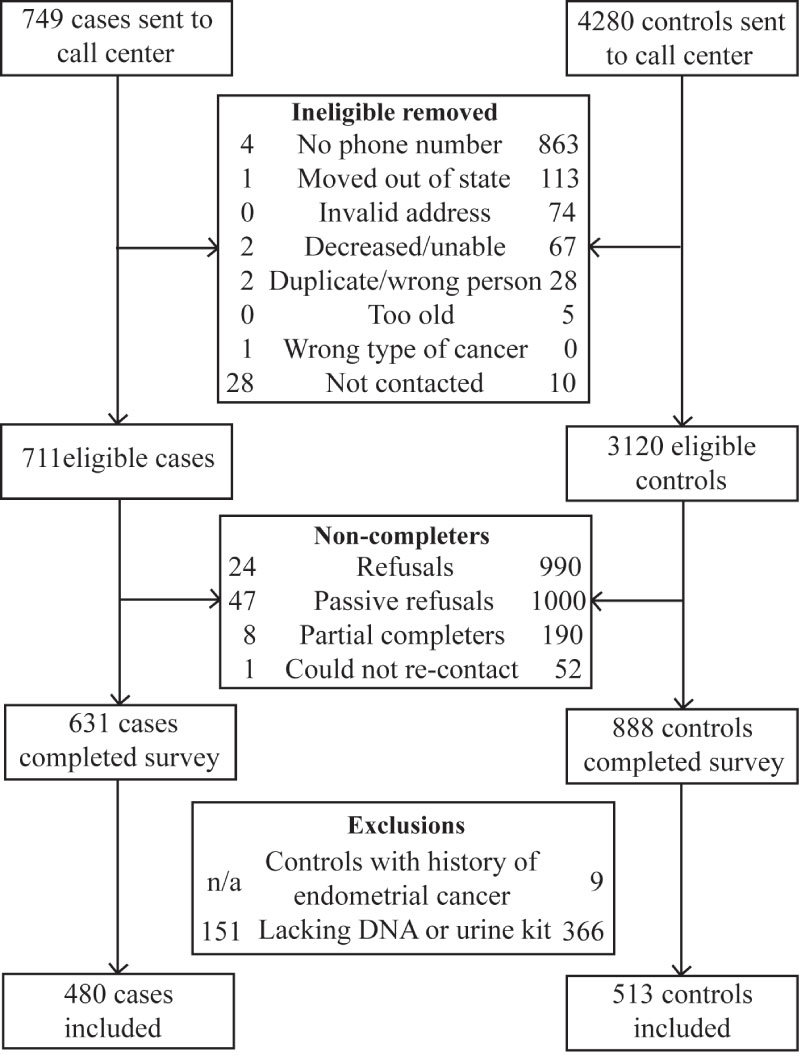 Figure 1: Exclusion and enrollment of participants in the Health and Environmental Exposure Research study.
View Figure 1
Figure 1: Exclusion and enrollment of participants in the Health and Environmental Exposure Research study.
View Figure 1
Endometrial cancer case participants and population-based controls were significantly different for all but 5 of the characteristics that we had selected a priori based on the literature (Table 1). These were smoking status, history of endometriosis, educational attainment, employment status and shift work. Among the commonly cited risk factors for endometrial cancer [13], case participants had higher average BMI (35.3 versus 29.0 kg/m2); a higher likelihood of diabetes (24.7% versus 12.7%), hypertension (57.5% versus 48.9%), first degree relatives with endometrial cancer (5.4% versus 1.2%), or ovarian cancer (6.3% versus 0.6%);menarche at age 11 years or earlier (25.7% versus 19.7%); and were more likely to be nulliparous (18.6% versus 11.5%) relative to controls. Among the 16 histology types identified from the cancer registry data, over 78% were endometroid carcinoma (ICD-03 code: 8380), 9% mixed cell adenocarcinoma (ICD-03 code: 8323), and 5% adenocarcinoma, NOS (ICD-03 code: 8140). The remaining histology types ranged from 1 to 12 cases, with 76 missing a histology type. Among the histology types with known histology 14% were Type 11 and 86% were Type 1. Since case ascertainment came directly from the cancer registries, the time of interview varied from 3.3 years since diagnosis to 4 months since diagnosis. Mean length of time from diagnosis was 1.6 years.
Table 1: Characteristics of endometrial cancer cases and population-based controls who returned urine and saliva samples*. View Table 1
Creatinine-adjusted urine cadmium levels varied from 0.006 to 0.649 (mean 0.041) μg/g in control subjects and from 0.005 to 0.417 (mean 0.037) μg/g in case participants. Urine cadmium concentration did not differ between cases and controls (P = 0.11). One SNP (rs4775936) and two of the ESR1 Haplotypes (CAC, TGT) were associated or nearly so with variation in creatinine-adjusted urine cadmium concentration (Table 2). For these loci, cadmium concentration was excluded from the conditional logistic regression model for case-control status since cadmium concentration made very little difference on parameter estimates.
Table 2: Association of alleles with creatinine-adjusted cadmium concentration. View Table 2
Eleven SNPs were successfully assayed from both DNA strands with a concordance rate among genotype pairs of 99.73%. Discordant genotypes were set to be missing values. Overall genotype call rate for the 11 SNPs was 98.35%. All SNPs were in Hardy-Weinberg Equilibrium (HWE; Bonferroni P > 0.08). In the single marker analyses, there were no genotype or allele frequency differences between the cases and controls (Table 3). Haplotype CAC at ESR1 (P = 0.01) and CG at AR (P = 0.05) were at a slightly higher frequency in the cases than in the controls (Table 3).
Table 3: Frequencies of participants' genotypes among endometrial cancer cases and population-based controls who returned urine and saliva samples. View Table 3
Of the 11 SNPs, the C allele at rs2234693 in ESR1 (P = 0.048) was associated with an increased risk of endometrial cancer in the conditional logistic regression multivariable models that fit the additive effect of SNP alleles on cancer risk (Table 4). Similarly, in the additive models for haplotype effects, carriers of the rare ESR1CAC haplotype (rs2234693:rs9340799:rs3020314) were associated with a significantly risk of endometrial cancer relative to carriers of all other haplotypes except TGC at ESR1 (Table 4). Marginal significance (p-value = 0.07) for endometrial cancer was observed for ESR2 GG haplotype (rs1255998:rs944050). In the non-additive model analyses, the dominance deviation d was not significantly different from 0 but the difference between alternate homozygotes was significant for rs2234693 in ESR1 (P = 0.039) (Table 5), consistent with an additive effect of this locus on risk of endometrial cancer revealed in the additive model analyses (Table 4).
Table 4: Association of loci or haplotypes with case-control status in multivariable logistic regression using additive coding. View Table 4
Table 5: Association of alleles with case-control status in multivariable logistic regression using non-additive coding*. View Table 5
Our study found suggestive evidence for associations between variants in ESR1 and ESR2 and risk for endometrial cancer. We found no evidence for associations between risk of endometrial cancer and variants in metallothionein 2A, a protein that sequesters cadmium, progesterone receptor, or androgen receptor. Our confidence in these finding is strengthened due to the extensive individual level data that were gathered and that allowed us to adjust the risk for known and suspected endometrial cancer risk factors.
Functionally, critical single nucleotide polymorphisms in many genes have been suggested as risk factors for the development of cancers such as endometrial cancer [5]. One consistently found and primary risk factor for endometrial cancer is estrogen exposure [14]. Previous studies that have evaluated the estrogen receptor 1 polymorphism rs2234693 in various populations (i.e., China, Sweden, Australia, Japan, Poland, USA, UK) with mixed results [6,15-26]. In a meta-analysis of thirteen studies, the estrogen receptor 1 polymorphism rs2234693 was significant for endometrial cancer risk (T vs. C allele: OR = 1.08, 95% CI, 1.00-1.17; and TT vs. CC allele: OR = 1.18, 95% CI, 1.00-1.38). In support of Zhou's finding, we also found an association between rs2234693 and endometrial cancer [27]. To our knowledge, only one other study has evaluated the estrogen receptor 2 polymorphism rs944050A/G allele for its effect on endometrial cancer risk and found an association [28]. In haplotype analyses of ESR1 (rs2234693:rs9340799:rs3020314) our study found an increased risk of endometrial cancer in women carrying the CAC haplotype. Finally, our study evaluated haplotype of ESR2 (rs1255998:rs944050) and found a marginally significant decreased risk of endometrial cancer in women carrying the GG haplotypes.
One limitation of our study was the very high proportion of white participants (> 95%), which prevented us from stratifying the analysis by race/ethnicity. Another potential source of ascertainment bias may have been caused by the fact that approximately one-third of the participants who contributed phenotypic data did not return saliva samples.
In conclusion, our study provides evidence that women carrying specific variants within estrogen receptor genes are at a slightly increased risk of endometrial cancer.
The authors acknowledge that the case data used in this report were provided by three cancer registries: The Arkansas Department of Health, Arkansas Central Cancer Registry (ACCR), Little Rock, AR; State Health Registry of Iowa, Iowa Cancer Registry (ICR), Iowa City Iowa; and the Missouri Cancer Registry and Research Center (MCR-ARC), Columbia, MO. Michele West and Jason Brubaker were key Iowa personnel involved in data collection for this project. Telephone interviews were conducted by the University of South Carolina's Survey Research Laboratory.
This work was supported by the American Cancer Society (RSG-10-197-01-CNE www.acs.org). Analytical instrumentation used in this study was acquired with support of the National Science Foundation, grant # BCS-0922374. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.