In previous studies, it has been reported that 10-20% of acute myeloid leukemia (AML) cases showed immunoglobulin heavy chain gene (IGH) rearrangements, a genetic hallmark of B-cell differentiation. However, the clinical significance of this is uncertain. Here, we report a case of diffuse large B-cell lymphoma (DLBCL) after complete remission (CR) from AML that exhibited an IGH rearrangement.
The patient was diagnosed with AML (M4Eo) with inversion of chromosome 16 [inv(16)]. Interestingly, the AML cells showed a monoallelic IGH rearrangement, as detected by the Southern blot analysis. The rearranged band was cloned using the inverse polymerase chain reaction (PCR) method. The sequence result revealed that a recombination between DH7-27 and JH4 occurred in the AML cells. PCR using DH7-27- and JH4-specific primers yielded no band from DLBCL specimens, suggesting that the DLBCL did not occur owing to the AML cell with IGH rearrangement. In addition, fluorescent in situ hybridization (FISH) showed that inv(16) was not found in the DLBCL cells. Therefore, although the AML cells harbored an IGH rearrangement, the origins of the two tumors seemed to differ from each other.
As the prognosis of AML became better, long-term follow-up studies of AML patients might define the clinical significance of IGH rearrangements in AML cells.
Acute myelogenous leukemia (M4Eo), Diffuse large B-Cell lymphoma, IGH rearrangement
Immunoglobulin heavy chain gene (IGH) rearrangement is a genetic hallmark of B-cell origin and differentiation [1-3]. However, it has been demonstrated in previous reports that IGH rearrangements were also found in approximately 10-20% of patients with acute myeloid leukemia (AML) [4-11]. However, the clinical characteristics of AML with IGH rearrangements remain to be identified.
AML after lymphoid malignancies is well described as therapy-related AML or myelodysplastic syndrome (MDS) [12,13]. These secondary myeloid malignancies are thought to be due to genetic mutations induced by chemotherapies or radiotherapies. However, the occurrence of diffuse large B-cell lymphoma (DLBCL) after myeloid neoplasms is not well described in the literature.
Here, we describe a rare case of DLBCL that occurred after complete remission (CR) of AML with inversion of chromosome 16 [inv(16)] (M4Eo). Interestingly, the AML cells exhibited IGH rearrangements, suggesting differentiation toward B-cells. Thus, the origin of the patient's AML an DLBCL cells was analyzed using IGH rearrangement and inv(16) as genetic markers.
A 54-year-old man was admitted to our hospital complaining from fever and dyspnea. There was no family history suggesting familial cancer syndromes or germline DNA repair mutations. Laboratory test results revealed anemia (hemoglobin: 8.5 g/dL; reference range: 13.5-17.5 g/dL), thrombocytopenia (platelet count: 4.5 × 109/L; reference range: 130-370 × 109/L), and elevated white blood cell count (11.9 × 109/L; reference range: 3.5-9.8 × 109/L) with 27% blasts. Bone marrow aspiration revealed a markedly variable number of immature monocytes and eosinocytes (Figure 1A). Leukemic cells were positive for CD13, CD33, CD34, and HLA-DR; and negative for CD19, CD7, and CD3 as measured by flow-cemetery. Inv(16) was detected using fluorescent in situ hybridization (FISH) (Figure 1B). Conventional karyotyping confirmed the inv(16) (data not shown). Based on the results, the patient was diagnosed with AML with eosinophilia (M4Eo). He achieved CR with conventional chemotherapy (induction chemotherapy and four courses of consolidation chemotherapy with anthracyclines and cytarabine). Then, he was followed up without maintenance chemotherapy at the outpatient clinic of our hospital.
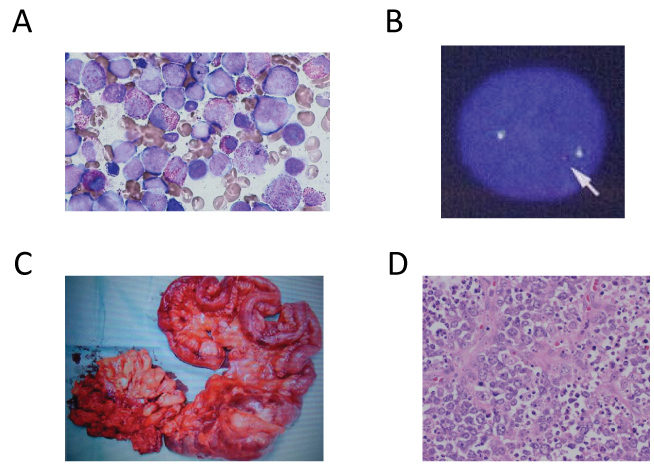 Figure 1: A) Bone marrow smear showing the proliferation of the blast, immature monocyte, and eosinocytes (May-Grünwald Giemsa staining, × 200); B) Interphase FISH for the CBFβ locus showing a split signal (arrow), indicating an inversion or translocation involving the occurrence of CBFβ; C) Resected ileocecal tumor; D) Ileocecal tumor showing diffuse proliferation of oval large lymphoid cells, which is consistent with DLBCL (hematoxylin and eosin staining, × 400).
View Figure 1
Figure 1: A) Bone marrow smear showing the proliferation of the blast, immature monocyte, and eosinocytes (May-Grünwald Giemsa staining, × 200); B) Interphase FISH for the CBFβ locus showing a split signal (arrow), indicating an inversion or translocation involving the occurrence of CBFβ; C) Resected ileocecal tumor; D) Ileocecal tumor showing diffuse proliferation of oval large lymphoid cells, which is consistent with DLBCL (hematoxylin and eosin staining, × 400).
View Figure 1
Three years after the diagnosis of AML, he presented at the emergency department of our hospital complaining of severe abdominal pain. He was diagnosed with peritonitis due to perforation of the ileocecal tumor (Figure 1C). The pathological findings of the ileocecal tumor revealed diffuse proliferation of the oval large lymphoid cells (Figure 1D). In addition, immuno histochemical staining demonstrated that the tumor cells were positive for CD10 and CD20 and negative for CD5, CD15, CD30, CD34, CD56, and cyclin D1. The tumor was diagnosed as DLBCL. He received four cycles of R-ACES regimen (Rituximab; 375 mg/m2 day1, Carboplatin; 100 mg/m2 day 2-5, Etoposide; 80 mg/m2 day2-5, methylprednisolone; 500 mg/body day2-6, Cytarabine; 2000 mg/m2 day6) for induction therapy. Then, he received R-MEAM regimen (Rituximab; 375 mg/m2 day-7, Ranimusutine; 250 mg/m2 day-7, Cytarabine; 400 mg/m2 day-6-3, Etoposide; 200 mg/m2 day-6-3, Melphalan; 140 mg/m2 day-2) as high dose chemotherapy followed by autologous peripheral blood stem-cell transplantation. He achieved CR, and then he was followed up at the outpatient clinic of our hospital without AML or DLBCL relapse.
Genomic DNA of the bone marrow cells in AML (M4Eo) at diagnosis was digested by PstI or HindIII. After electrophoresis in 0.8% agarose gel and 1 × TAE buffer, the digested DNA was transferred onto a nylon membrane (Roche, Mannheim, Germany). A DNA fragment of the IGH joining region was labeled with digoxigenin using a DIG-DNA labeling kit (Roche) and used as a probe according to the manufacturer's protocol. Hybridization, washing, and detection were also performed according to the manufacturer's protocols (Roche).
In order to determine the nucleotide alignment of the IGH rearranged band seen in Southern blot analysis, inverse PCR was performed to clone the rearranged band using stocked genomic DNA obtained at the onset of AML(M4Eo), as reported elsewhere [14]. Briefly, 1 μg of genomic DNA of the AML cells was digested by PstI completely. The aliquot of 10 ng/mL of digested DNA was reacted with T4 ligase at 4 °C for 16 h, in order to generate self-ligated circular DNA. The circular DNA was then subjected to nested PCR using LA-Taq polymerase (Takara, Shiga, Japan). After electrophoresis, the PCR product of interest was isolated using a gel extraction kit (QIAGEN, Hilden, Germany). The nucleotide alignment of the PCR product was determined directly using Sanger's method.
The forward (5'-TGAGCTGAGAACCACTGTGC-3') and reverse (5'-AGGAGACCCAGCACCCTTAT-3') primers were set at 5'-upstream DH and 3'-downstream JH4, respectively. The estimated PCR products of IGH rearrangement of AML (M4Eo) and germ line of control cells were calculated as 177 bp and 1,448 bp, respectively. The genomic DNA of the DLBCL was extracted from a paraffin-embedded tissue piece according to the manufacturer's protocol by TaKaRa DEXPAT (Takara, Kyoto Japan). PCR was performed on the patient's DLBCL and AML cells using the primers.
FISH was performed on the interphase nuclei using CBFβ split probes set at 5' and 3' of the CBFβ locus. When an inversion or translocation occurred around the CBFβ locus, the probes showed a split signal. FISH was performed at another institute (BML, Kawagoe, Japan).
Possible IGH rearrangements in the AML (M4Eo) cells before the onset of DLBCL were assessed. Interestingly, the Southern blot analysis of the AML cells showed a monoallelic IGH rearranged band (Figure 2A) in various endonuclease digestions. The 2.4 kb rearranged band found in PstI digestion was amplified and cloned using inverse-PCR (Figure 2B). The nucleotide alignment of the PCR product revealed that the AML cells underwent recombination between DH7-27 and JH4 (Figure 2C).
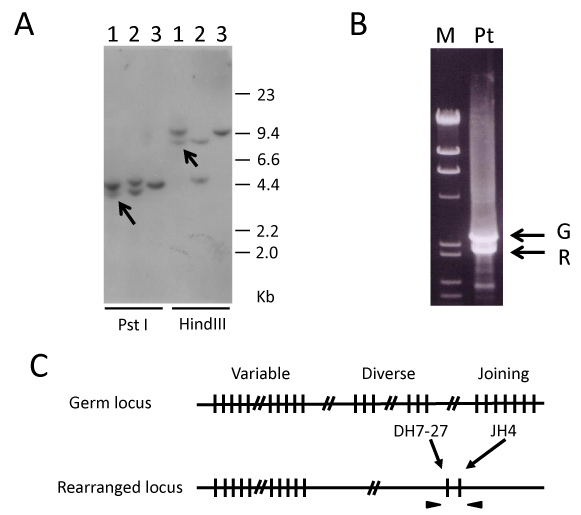 Figure 2: A) Southern blot analysis of the IGH-JH region. Line 1: Bone marrow cells obtained at AML diagnosis (M4Eo). Line 2: Positive control (B-cell lymphoma cell line, KHM2B). Line 3: Negative control (myeloid tumor cell line, HL60). The arrows indicate rearranged bands. All DNA was digested by PstI or HindIII; B) Molecular cloning of the rearranged IGH-JH region by inverse PCR using self-ligated circular DNA after PstI digestion as a template. M: DNA marker (λ/HindIII+φ/HaeIII); Pt: Bone marrow cells obtained at AML diagnosis (M4Eo). The upper and lower arrows indicate germ line (G) and rearranged (R) bands, respectively; C) AML (M4Eo) cells showing a recombination between DH7-27 and JH4. The arrowheads represent primers amplifying the recombination DNA.
View Figure 2
Figure 2: A) Southern blot analysis of the IGH-JH region. Line 1: Bone marrow cells obtained at AML diagnosis (M4Eo). Line 2: Positive control (B-cell lymphoma cell line, KHM2B). Line 3: Negative control (myeloid tumor cell line, HL60). The arrows indicate rearranged bands. All DNA was digested by PstI or HindIII; B) Molecular cloning of the rearranged IGH-JH region by inverse PCR using self-ligated circular DNA after PstI digestion as a template. M: DNA marker (λ/HindIII+φ/HaeIII); Pt: Bone marrow cells obtained at AML diagnosis (M4Eo). The upper and lower arrows indicate germ line (G) and rearranged (R) bands, respectively; C) AML (M4Eo) cells showing a recombination between DH7-27 and JH4. The arrowheads represent primers amplifying the recombination DNA.
View Figure 2
Furthermore, whether the clonal IGH rearrangement of the AML cells was found in the DLBCL cells was assessed. We prepared specific primers that can amplify the recombined sequence (DH7-27 and JH4) of the rearranged IGH locus of the AML cells. The expected band (177 bp) was detected in AML (M4Eo); however, PCR of this patient's DLBCL cells was performed using the primers, no band was detected (Figure 3A). Thus, this result suggested that IGH rearrangements in DLBCL cells differ from those in AML cells.
 Figure 3: A) 1,448 bp DNA representing the DH/JH4 germ line amplified in the germ line control DNA (lane 1). A 177 bp DH/JH4 recombination fragment amplified in the AML (M4Eo) sample (lane 2, arrow). Lane 1: HL60 (germ line control). Lane 2: AML DNA (M4Eo). Lanes 3 and 4: DLBCL DNA. Lane 5: Negative control (H2O); B) FISH for inv(16) in DLBCL cells showing no split signal.
View Figure 3
Figure 3: A) 1,448 bp DNA representing the DH/JH4 germ line amplified in the germ line control DNA (lane 1). A 177 bp DH/JH4 recombination fragment amplified in the AML (M4Eo) sample (lane 2, arrow). Lane 1: HL60 (germ line control). Lane 2: AML DNA (M4Eo). Lanes 3 and 4: DLBCL DNA. Lane 5: Negative control (H2O); B) FISH for inv(16) in DLBCL cells showing no split signal.
View Figure 3
Inv(16) in this patient's DLBCL cells was analyzed using FISH. The DLBCL cells showed no split signal, indicating that they did not harbor inv(16) (Figure 3B).
Here, we reported a patient who developed secondary DLBCL during the CR of AML. Interestingly, the AML cells harbored an IGH rearrangement and inv(16). Therefore, the tumor origins of AML and DLBCL were examined by genetic analyses. As a result, the origins were found to be different from each other in terms of IGH rearrangement and inv(16). However, we did not rule out that some other genomic mutations might be predisposed to the development of the both hematological malignancies.
Myeloid malignancies after chemotherapy or radiotherapy are well known as therapy-related AML or MDS [12,13]. However, secondary lymphoid tumors following AML are rare. Higuchi, et al. reported Epstein-Barr virus-positive DLBCL during CR of AML [15]. In this case, the rearrangement patterns of the IGH in the two malignancies were different. However, they showed that these two malignancies had a common clonal origin, indicated by chromosomal translocation t(3;4) (p25;q21); therefore, they concluded that the tumors possibly had common origins. In our case, the tumor origins of AML and DLBCL cells were found to be different from each other, according to the genetic analyses of IGH rearrangement and inv(16).
It has been shown in previous reports that AML exhibited an IGH gene rearrangement in approximately 10-20% of all cases. However, the clinical features of AML with IGH rearrangement are still unidentified. Kyoda, et al. reported that IGH-rearrangements of AML may be considered as a poor prognostic factor of CR and survival [6]. However, it has been shown in other reports that IGH rearrangement of AML did not affect the prognosis [7]. These differences may be due to the short observation period. Recently, there has been an improvement in the survival of patients with AML [16]. Therefore, the characteristics and clinical features of AML with IGH rearrangement might become clear in a long-term observation period.
In conclusion, we reported a rare case of DLBCL that developed during the CR of AML with IGH rearrangement and inv(16). Since IGH rearrangements due to clonal B cell proliferation can be found among older persons, IGH rearrangements might be just coincidence in AML. Further studies are required to determine the clinical significance of IGH rearrangements in AML.
All authors declare to have no conflicts of interest in this work.