This study investigated the antibiogram profile of Serratia marcescens among hospitalized individuals, hospital environments and halls of residence of Obafemi Awolowo University in Ile Ife, Osun state with a view to provide key information on resistance factors that are of therapeutic importance among the understudied pathogen.
Two hundred and twenty samples from clinical and non-clinical sources were collected with ethical clearance approval (ERC/2018/09/02) from the hospital advisory committee. They were cultured on sorbitol Mcconkey agar infused with 200 U/ml of colistin for the selective isolation of Serratia species. Non-duplicate colony was picked from each cultures and characterized biochemically with the use of microbat 24E kit to identify various species isolated. The susceptibility profiles of the isolates against selected antibiotics were studied by the Kirby-baeur disc-diffusion technique. Results were interpreted according to CLSI (2019).
Thirteen (61.9%) Serratia marcescens out of the 21 (9.55%) Serratia sp. isolates were recovered from the study area. Other species isolated were S. fonticola (4.8%), S. rubidae (4.8%) and Serratia liquefaciens complex (28.6%). Serratia species were significantly associated with the community (OAU) than the hospital (OAUTHC) (p ˂ 0.001). Serratia marcescens showed great susceptibility to meropenem (100%), ofloxacin (100%), ciprofloxacin (90.5%) and gentamicin (84.6%), and total (100%) resistance to ceftazidime, cefuroxime, ampicillin and augmentin.
The study concluded that occurrence of multidrug resistant S. marcescens pose a public health threat in the study area.
The frequency of infections and outbreaks due to S. marcescens has steadily increased for 4 decades, especially among infants, the hospitalized and individuals with suboptimal immunity [1]. Amidst such individuals, unsuccessful antibiotic therapy due to resistance results sometimes to death, permanent organ damage and more often; longer course of infections.
Antibiotic resistance among Gram negative bacteria is a public health problem of global relevance; resistance among Serratia sp. are more worrisome due to their enormous virulence factors, ability to exist in biofilm resulting in polymicrobial infections, chromosomal resistance factors e.g. enzymes and genes as well as those that could be acquired as plasmids [2]. Aminoglycosides resistance is common among Serratia sp. [3].
Serratia marcescens accounts for most infections caused by members of the Serratia genus and have been recovered from various clinical specimens [4]. S. marcescens has been incriminated in urinary tract infections, meningitis, pneumonia, septicemia, endocarditis, arthritis, and wound infections [4]. Serratia marcescens has now been regarded as a pathogen in every conceivable kind of infections [5] however; the presence of underlying immunosuppressive or debilitating conditions is critical to establishment of infection among the hosts [6]. These conditions may include heroin addiction [5], ageing [7] prolonged antibiotic therapy or hospitalization with indwelling devices like catheter, oxygen aid, blood bags, ultrasonic nebulizers, bronchoscope [1], HIV/AIDS [8], or poor immune development levels among neonates and babies [9].
Kisirak and Hukic [10] reported an outbreak of post-surgical Serratia marcescens infections involving 79 cases within 2010 and 2011 in the orthopaedic clinic of university of Sarajevo. Isolates were recovered from wound swabs, blood cultures and cerebrospinal fluid of patients and bottles of anesthesia used in course of surgeries of the cases earlier on. The anesthesia bottle was incriminated as the reservoir of the outbreak strain following the successful isolation from them. The isolates were multi drug resistant but sensitive to tested carbapenem in their studies. A case of fatal meningitis due to S. marcescens was earlier recorded in the same hospital in 2007 that implicated anesthesia bottle as reservoir for the pathogen [11].
Outbreaks of nosocomial infections caused by Serratia marcescens especially among immunosuppressed individuals have been reported around the world. In Ile Ife, the limited information on S. marcescens resistance profile present patients with risk of longer course of infection and treatment, hence this study.
A total of 220 samples were collected from hospitalized patients of Obafemi Awolowo university teaching hospitals, hospital and hall environments of Obafemi Awolowo University within October 1 to December 30, 2018 with an approval (ERC/2018/09/02) by the medical advisory committee. Samples collected were urine from catheterized patients, swabs of surfaces, door handles, sinks and soap containers from selected wards. Swabbed samples of the university hall environments including bathroom tiles, and door handles were also collected following standard microbiological methods [12]. Urine samples were collected in sterile universal bottle and transported to the laboratory. Each swabbed sample was transported in bijou bottles containing 5 ml of sterile tryptic soy broth. All samples were transferred to the laboratory for immediate cultures on freshly prepared sorbitol MacConkey agar infused with 200 U/ml of colistin.
All samples were inoculated by spread plate method on Sorbitol MacConkey Agar (SMAC; Oxoid CM0813; Hampshire, England) infused with Polymyxin B (colistin) (200 U/ml) as described with slight modification. Following manufacturer's recommendation, sorbitol MacConkey agar powder were dissolved in sterile distilled water (5.15 g/100 ml) in an air-tight conical flask. The mixture was homogenized and autoclaved at 121 °C for 15 minutes. The molten agar was allowed to cool to 45 °C prior to addition of dissolved colistin. Vials of Polymyxin B powder (Alvogen; New Jersey, USA) 500,000 U were dissolved in sterile distilled water and added to sterile, molten but cooled SMAC to make 200 U/ml concentration. Each conical flask was shaken to ensure even distribution of colistin in the molten agar, and poured into sterile petri dishes. The agar plates were allowed to gel, and dried in laboratory oven (45 °C) to remove condensed water vapors. Each sample was inoculated by spread plate method. The inoculated agar plates were incubated at 28 ± 2 °C for 24 hours to enhance pigmentation of Serratia sp. [5]. A colony of pink/red sorbitol/non-fermenting pigmented or non-pigmented colony was picked per plate. Pure colonies of each isolate were obtained by sub culturing each isolate on freshly prepared nutrient agar prior to biochemical characterization for identification.
Cultural, biochemical characterization and Identification of Serratia sp. were carried out following procedures of [12]. Standard procedures on Gram staining, motility test, oxidase test, gelatin hydrolytic test, DNAse and Casein Hydrolysis were conducted.
Further identification of isolates was done using Microbact 24E identification (Basingstoke, England) which is a standardized micro-substrate system for the identification of Enterobacteriaceae and common miscellaneous Gram-negative bacilli. Each kit of 24 miniature biochemical tests identifies GNB to species with proven degree of accuracy. Organism identification is based on pH change and substrate utilization. Each well was inoculated with an isolate suspension that reconstitutes the media. During incubation at 37 °C for 24 hours and 48 hours, metabolic reactions produced colour changes that were either spontaneous or revealed by the addition of reagents. The results were recorded on report forms and interpreted using the Microbact Identification Package. E. coli ATCC 25922 served as the control strain.
Two to three isolated colonies from the 18-24 hours cultures were emulsified in 5 mls of normal saline; it was mixed thoroughly to prepare a homogenous suspension.
The microbact test kit used was 24E. With the aid of sterile micro pipette and sterile tips 100 μl of the homogenous suspension were added to each test strips. The wells lysine, ornithine, hydrogen sulphide, containing the homogenous suspension were overlaid with mineral oil. The strips were incubated for 24 hours and 48 hours at 37 °C. The results were read after adding necessary reagents such as indole, VP and TDA.
This test was performed in well 7 (ONPG) after reading the ONPG reaction. One drop of Nitrate Reagent A and 1 drop of Nitrate Reagent B were added to the well.
The 24E strip was read after 24 hours and the reactions were evaluated as "positive" or "negative" by comparing to the colour chart and recording the results. The following reagents were added; Well 8 (Indole production), 2 drops of indole (Kovacs) reagent was added and was evaluated within 2 minutes of the addition of the reagents. Well 10 (Voges-Proskaüer-reaction), one drop each of VPI and VPII reagent was added and evaluation was done 15-30 minutes after the addition of the reagents. Well 12 (Tryptophan deaminase), one drop of TDA reagent was added and the test was evaluated immediately after the addition of the reagent.
An octal coding system was adopted for Microbact. Each group of three reactions produces a single digit of the code. Each digit of the code is formed by adding the indices of the positive reactions. A total of eight digits from the eight groups form each code number (octal code). These codes were entered into the software package from microbact. Percentage probabilities were recorded along with isolate IDs.
The antibiotic sensitivity profile of each isolates against selected antibiotics was performed by the Kirby Bauer Disc diffusion technique as described by Cheesebrough [12] and interpreted following the Clinical Laboratory Standards Institute [13]. The isolates were inoculated on Muller Hinton agar (Oxoid, USA). Overnight culture of each isolate was adjusted to 0.5 McFarland standard in 0.85% sterile normal saline. Antibiotic disks used in this study include: Ceftazidime; 30 μg, Cufuroxime; 30 μg, Gentamicin; 10 μg, Ciprofloxacin; 5 μg, Ofloxacin; 5 μg, and Amoxicillin/Clavulanate; 30 μg, Nitrofurantoin; 300 μg, Ampicillin; 10 μg, Aztreonam (10 μg) and Cabapenems: Imipenem; 10 μg, and Meropenem; 10 μg all supplied by Oxoid, USA. The diameters of zone of inhibitions of each antibiotics were measured and interpreted following the Clinical Laboratory Standards Institute (2019) [13] interpretative charts. Results were recorded as sensitive 'S' and Resistant 'R' following CLSI (2019) recommendations.
Data obtained were analyzed and presented as frequencies and percentages. Data were compared with the use of the two tailed Fishers Exact Test with the SPSS statistical program (version 22). All reported p-values were two-sided and a p-value of less than or equal to 0.05 was considered to be statistically significant.
The results showed 9.55% prevalence of Serratia isolates in the study population where two hundred and twenty (220) non-duplicate consecutive samples were collected from hospitalized patients Obafemi Awolowo University Teaching Hospital Complex (OAUTHC), hospital environments and hall environments of the University. Serratia species are significantly associated with the community (OAU) than the hospital (p ˂ 0.001).
The Serratia species were isolated from the diverse range of specimens between October and December 2018. All 21 (100%) Serratia species were isolated from the environment and none (0%) from humans (Table 1).
Table 1: Isolated Serratia sp. and their source. View Table 1
The recovered Serratia species (n = 21) exhibited high antimicrobial susceptibility to ofloxacin 21/21 (100%), meropenem 21/21 (100%), ciprofloxacin 19/21 (90.5%), gentamicin 18/21 (85.7%), imipenem 14/21 (66.6%) and showed low susceptibility to Nitrofurantoin 9/21 (42.9%). There was no susceptibility to ceftazidime (0%), cefuroxime (0%), ampicillin (0%) and augmentin (0%) as represented in Figure 1.
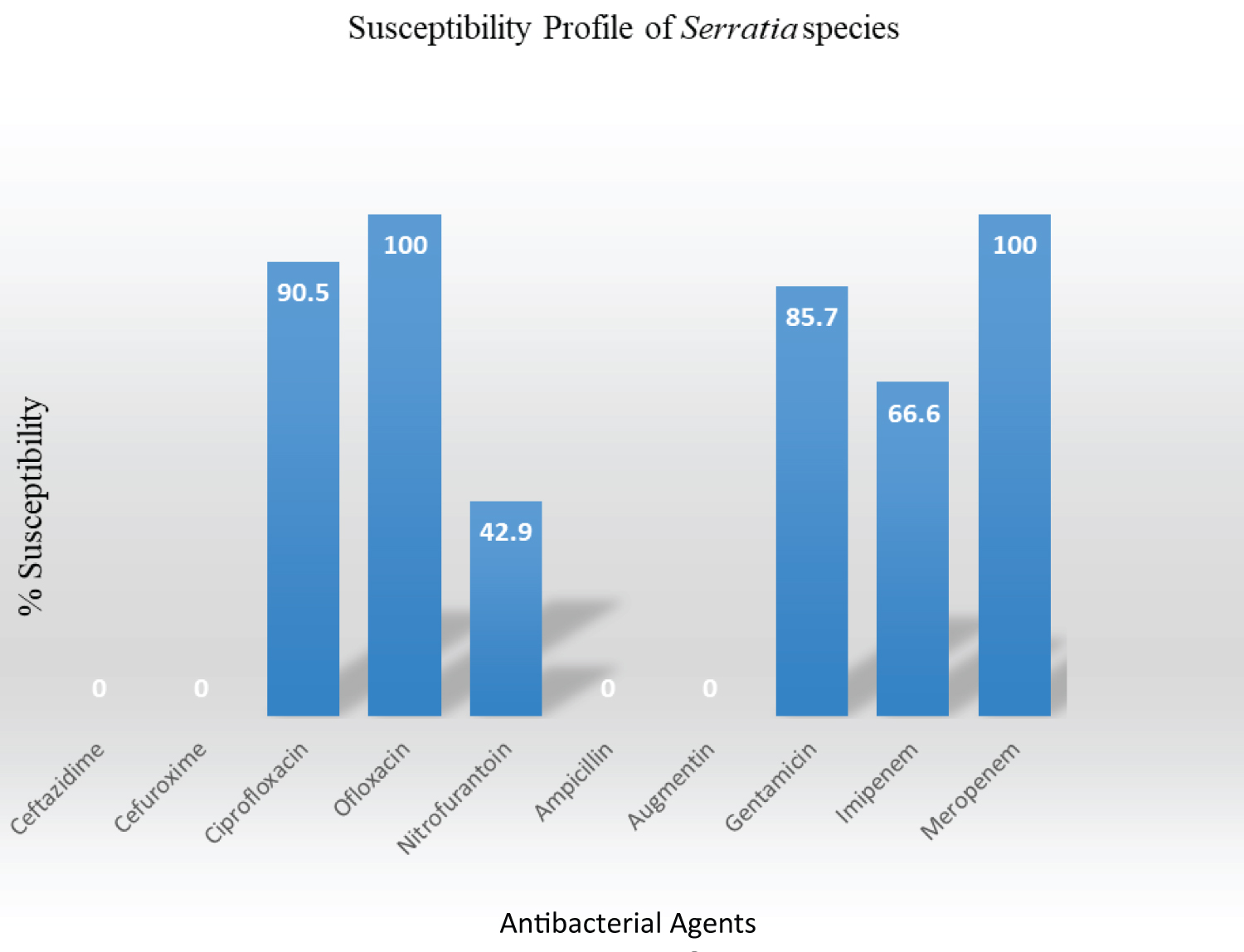 Figure 1: Antibiogram profile of isolates. View Figure 1
Figure 1: Antibiogram profile of isolates. View Figure 1
S. marcescens showed great susceptibility to meropenem (100%), azithromycin (100%), ofloxacin (100%), ciprofloxacin (90.5%) and gentamicin (84.6%). S. fonticola was susceptibility to nitrofurantoin (100%), meropenem (100%), azithromycin (100%), ofloxacin (100%) and gentamicin (100%). S. rubidaea was resistant to ceftazidime (100%), cefuroxime (100%), ampicillin (100%) and augmentin (100%) from the tested antibiotics as shown in Table 2.
Table 2: Relative susceptibility patterns of Serratia spp. View Table 2
Increasing antibiotic resistance remain a global public health concern, with particularly serious consequences in developing countries like Nigeria, where resistance has dramatic effects on morbidity and mortality rates and threatens the viability of local healthcare systems.
All the isolated Serratia species in this study demonstrated very poor susceptibility patterns to most of the antimicrobials from major antibiotics classes tested using CLSI (2019) breakpoint. This is a very alarming trend and predicts that infections caused by S. marcescens may be difficult to treat.
There have been reports regarding the identification, antibiotic susceptibility, pathogenicity and epidemiological investigation of this microorganism [5]. The outbreaks of Serratia species have been described; and reservoirs of the pathogens identified in a variety of clinical settings and different environmental sources that makes accurate identification of Serratia species important in defining outbreaks [14]. In the study area, there is paucity of data on the antibiotics susceptibility profile of this group of organisms; which are of epidemiological and therapeutic importance for the locality and the nation as a whole.
Serratia marcescens in this study were only among others highly susceptible to ofloxacin, meropenem, ciprofloxacin in agreement with the report of Al Jarousha, et al. [15] which indicated that meropenem, ciprofloxacin and ofloxacin were most effective in the treatment of S. marcescens infections with 90%, 76% and 73% success respectively.
There is variation in the susceptibility pattern of the Serratia species isolated in this study; S. marcescens were susceptible to meropenem, azithromycin, ofloxacin, while the S. fonticola was susceptibility to ofloxacin, gentamicin, nitrofurantoin, meropenem and azithromycin. This is in line report of Mahlen [3], where some strains of Serratia were sensitive to cefuroxime and resistant to Ceftazidime, some could be resistant to tobramycin and yet be resistant to gentamicin, this led Mahlen (2011) [3] to recommend that every Serratia infection treatment should be preceded by a laboratory sensitivity test.
Almost all antibiotics, except the tested fluoroquinolones and carbapenems were generally ineffective against Serratia marcescens in this study. To control the emergence and spread of the new multidrug-resistant Serratia species, more research should targeted on alternative pathogens control techniques as quorum quenching.
ABO planned and performed the experiment while JOA Supervised and Coordinated all experiment and wrote the manuscript with contributions from both Authors.
We are grateful to Dr. Folashade Osotimein of Albany State University, New York for her technical advice and reagents supplied in part for the successful completion of the study.