Serious gastrointestinal infection of Rotavirus (RV) is usually prevalent during winter months and often seen in infants and young children. Studies on genotypes of prevalent rotavirus strains are quite important for preventing infection, developing vaccines, and its evaluation. While at least 11 G genotypes have been isolated from humans, G1, G2, G3, G4, and emerging G9 are major genotypes of human RV. Although G6 human rotavirus is quite rare, it is the major type among rotaviruses from cattle. In this study we investigated the characteristics and origin of bovine RV strains from the children of the Nasu district, Tochigi, Japan.
We examined the clinical findings in 147 patients who attended to the Department of Pediatrics at International University of Health and Welfare Hospital in Nasu-shiobara City, Tochigi, Japan during the time of April 1, 2008 to March 31, 2010. We analyzed the clinical findings of the 37 patients with a fecal sample positive for RV antigen on VP6, NSP4 gene RT-PCR genotyping. Viral genotypes were determined using rotavirus-positive samples from 27 of these 37 patients.
In 37 cases in VP6, NSP4 gene-based RT-PCR 24 samples were positive and the genotypes were determined as G1P[8] in 5 samples, G3P[8] in 5 samples, G9P[8] in 3 samples, and G6P[9] in 2 samples. Of particular note, we detected G6P[9] which were extremely rare in human beings but common in cattle. KF17 strain of a patient showed G6 type of bovine strains and P[9] was admitted to reconcile with human strains. Per match ratio in each gene of VP6, VP1, VP2, VP3, and NSP4 showed a high match rate (84.1-86.4%) with DS-1 type. Per match ratio in each gene of VP4, NSP1, NSP3, NSP4 and NSP5 also showed a high match rate (94.2-96.9%) with AU-1 type. Bovine type strain gene segments become reassortant rotaviruses major two genes in these stocks.
In the phylogenetic analysis the gene of a strain KF17 of G6P[9] was located in a human lineage including other human G6 strains. Similarly, all other genes of the strain except for the NSP3 gene were relatively closely related to at least one of the human G6 RVs reported in Europe and the U.S. In a study in Miyagi Prefecture G6P[9] (M72S11) per sample was found from a 2-year-old toddler in 2011. Also G6P[9] have been detected from more than 3 cats of Mie Prefecture. These findings suggest that human G6 RVs which had occurred by reassortment between human and bovine RVs are distributed worldwide, despite low prevalence. G6 RV may occur independently in different areas through reassortment among local strains.
Rotavirus gastroenteritis, Endemic strain, Genotype, G6P[9], Bovine rotavirus, Reassortment
Rotavirus (RV), is an important viral pathogen causing Acute Gastroenteritis (AGE) in humans. As most of infants have received the rotavirus vaccine rotavirus infections among infants and young children has since decreased significantly in developed countries [1]. Each year, the vaccine prevents an estimated 40,000 to 50,000 hospitalizations among U.S. infants and young children [2]. RV infects not only humans but non-human monkeys and bovine, pigs, horses, dogs, cats, rats and chicken. Nature of RV in spans of many mammals and birds, infection of rotavirus infection in humans is virtually limited to humans. Reports investigated the epidemic strain in local areas are very important to investigate the pandemic from the epidemic in the region.
RV, a member of the family Reoviridae, has 11 segments of double-stranded RNA as a genome, and the viral particle is composed of the outer capsid, inner capsid, and core [3]. The outer capsid consists of two structural proteins, VP4 and VP7, which contain neutralization antigens. The inner capsid consists of structural protein VP6. Based on the antigenicity of the inner capsid protein VP6 and genomic characteristics, rotavirus is classified into seven groups (A-G), among which group A RV is the major etiologic agent in humans and animals. For epidemiological investigations of RV, a genetic classification system based on the outer capsid proteins VP7 (G type) and VP4 (P type) has been adopted [4]. While at least 11 G genotypes have been isolated from humans, G1, G2, G3, G4, and emerging G9 are major genotypes of human rotaviruses. As human P genotypes, P[8] is the most common genotype worldwide, followed by P[4] and P[6]. Also, 6 Non-Structural Proteins (NSP) are known.
In human RVs, five common G and P genotype combinations (genogroups) have been identified: G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8] on the Wa-like genome constellation (I1-R1-C1-M1-A1-N1-T1-E1-H1), and G2P[4] on the DS-1-like constellation (I2-R2-C2-M2-A2-N2-T2-E2-H2) [3]. It was considered that RV infection in humans is virtually limited to humans. AU-1-like rotaviruses make up a third group of human RV, which is distributed at low prevalence and has a distinct gene constellation (G3-P[9]-I3-R3-C3-M3-A3-N3-T3-E3-H3). So far, few G6P[9] rotaviruses have been detected in humans. The first G6P[9] strain, PA151, was isolated from an Italian child with AGE, followed by the Se584 strain from the United States and several Hungarian strains [5]. Although G6 human rotavirus is quite rare, it is the major type among rotaviruses from cattle. In a study of Japanese cows, 59.1% of isolates belonged to G6. Usually, bovine G6 strains were combined with P[5], P[1], and P[11] worldwide [6,7].
In Japan, Rotarix and RotaTeq have been on the market since November 2011 and July 2012, respectively. Although RV vaccination had not been adopted into the national immunization program as of 2014, an estimation of 45% uptake with a wide range of variation throughout Japan was reported in 2013. The number of infectious gastroenteritis cases including RV-AGE has been collected weekly from sentinel pediatric clinics since the years before vaccine introduction.
Recently, full-genome sequences of rotavirus strains have been increasingly analyzed in order to understand the interspecies transmission, reassortment, and evolutionary relationships between human and animal rotaviruses. In the previous study [8,9], nearly full-length sequences of all the gene segments were determined to investigate the genetic origin of the unique human G6P[9] rotaviruses detected recently in Japan. In this study we investigated the characteristics and origin of RV strains from the children of the Nasu district, Tochigi, Japan.
During the time of April 1, 2008 to Mar 31, 2010 children less than 15 years of age who suffered from fever, diarrhea, vomiting, abdominal pain, consulted the Department of Pediatrics, International University of Health and Welfare Hospital, Nasu-shiobara, Tochigi, Japan were investigated for clinical and laboratory findings, infectious disease epidemic situations and past medical histories. Considering the findings of 147 patients diagnosed with AGE, referring to the results from stool culture and simple rapid tests using immunochromatography (Sekisui Medical Co., Tokyo, Japan) of stool samples for RV. Swabs of stool specimen and cerebrospinal fluid and blood specimens were collected and kept at -20 ℃. None of the147 children had traveled abroad. Twenty-three patients out of 37 rapid tests using immunochromatography rotavirus antigens-positive admitted to the hospital.
Genetic analysis was performed with 24 fecal, 1 cerebrospinal and 2 blood specimens. Presence of RV antigens in stool specimens was confirmed with immunochromatography. All specimens were stored at -80 ℃ until analyzed. G types [1-4, 8, 9, 12], P types ([4], [6], [8], [9]), VP6, and NSP4 genotypes were determined by the nested PCR with the primer sets reported previously [10-12].
Nucleotide sequences were determined directly with the cDNA products amplified by RT-PCR. As a template for RT-PCR, dsRNA was extracted from stool suspension with a commercially available kit (RNAID kit, BIO101, Inc., La Jolla, CA, USA) according to the manufacturer's instructions. RT-PCR was performed with reverse transcriptase (AMV) (Seikagaku Co., Tokyo, Japan) and thermostable DNA polymerase (Expanded High Fidelity PCR System, Roche, Mannheim, Germany) with the primers for individual gene segments prepared based on the human rotavirus strains reported previously [8,9]. For all the gene segments, full-length sequences except for primer binding regions at 50- and 30-end were amplified and sequenced. PCR products were purified by Wizard® SV GEL and PCR Clean-Up System (Promega, Inc., Madison, WI). Sequencing reaction was performed with fluorescent dideoxy chain termination chemistry using the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA).
Sequence was determined by ABI Prism 3100 genetic analyzer (Applied Biosystems). GENETYX-Win version 5.1 (Software Development, Tokyo, Japan) was used to calculate the identity of gene segments. Phylogenetic analysis was performed with MEGA software version 4.1 based on the neighbor-joining method and the Kimura two-parameter model. Phylogenetic trees were supported statistically by bootstrapping with 1,000 replicates. The nucleotide sequences of 11 gene segments of the KF17 strain determined in this study were deposited in the GenBank database under accession numbers JF421975-JF421985 [13].
Specimen collection was performed by the authors' responsibilities under the agreement based on ethical guidelines on clinical research (issued by the Ministry of Education and Science and the Ministry of Health, Welfare and Labor of Japan) and the Helsinki Declaration (World Medical Association). The study plan was approved by the Research Ethics Committee of International University of Health and Welfare.
Age distribution of patients was from 7 months to 2 years. RV infections were mainly distributed to the age of 7 months to 2 years and adenovirus infections in 7 to 11 months. RV was detected in 27 specimens examined by RT-PCR, although three NSP4 genotypes were positive (21 B, 1 C, 1 E3) (Table 1).
Table 1: Summary of genetic analysis of rotavirus strains in Japan. View Table 1
Subsequent sequence analysis revealed that this genotype E3 (KF17) strain had the G6-VP7 gene which is rarely found in human rotavirus, and the full-length VP7 gene sequence and partial VP4 gene sequence of the KF17 were preformed (Figure 1). Therefore, nearly full-length sequences of all 11 RNA segments were determined for the KF17 strain. Strains KF17 were obtained from 3-year-old female outpatient (Case 24) in February 2010. Two G6-positive patients (Cases 24 and 25) were from different families, and lived in the same Tochigi prefecture town, one which has plenty of cattle farms. However, there was no evidence that they were in contact with each other, or with cattle or other animals. Trisomy 13 had been diagnosed for the Case 24.
 Figure 1: Phylogenetic trees of the VP7 gene segments of the KF17 strain from Case 24.
View Figure 1
Figure 1: Phylogenetic trees of the VP7 gene segments of the KF17 strain from Case 24.
View Figure 1
Out of the 15 RV strains which were determined for their G or P types; two strains were identified as P[9] and 13 as P[8]. Strains were determined as G1 (five specimens), G3 (five specimen) and G9 (three specimens). Immune electrophoresis results showed G genes (VP7) in Cases 2, 3, 21, 22, 16 were G1 (618 bp), Cases 4, 5, 9 were G9 (179 bp) and Cases 7, 16, 17, 18, 23 were G3 (682 bp). The P gene (VP4) was determined as [8] (345 bp) in all cases of 2-5, 7, 9, 16-18, 21-26. VP6 genotype was determined as II (485 bp) in all cases of 2-5, 7, 9-20, 22-24, 26, 27. NSP4 genes in Cases 2-4, 7, 9-20, 22, 23, 26, 27 were B (618 bp), Cases 24 was E3 and Case 25 was C. The genotypes of the current classification are now in progress.
By the phylogenetic analysis 2 samples were determined as G6P[9]. Especially G6P[9] per 11 gene phylogenetic analysis available was performed. VP7 gene of Case 5 G9P[8] was located in single lineage extends to the world. VP7 gene of Case 24 G6P[9] (KF17 strain) was partially located closer to the cattle stock. Phylogenetic analysis of KF17 was performed on VP4, VP6, VP1, VP2, VP3, NSP1, NSP2, NSP3, NSP4 and NSP5 genes and determined as G6P[8], I2-R2-C2-M2-A3-N2-T3-E3-H3. KF17 strain of Case 24 showed G6 type of bovine strains and P[9] was admitted to reconcile with human strains (Table 2). Per match ratio in each gene of VP6, VP1, VP2, VP3, and NSP4 showed a high match rate (84.1-86.4%) with DS-1 type. Per match ratio in each gene of VP4, NSP1, NSP3, NSP4 and NSP5 also showed a high match rate (94.2-96.9%) with AU-1 type. Bovine type strain gene segments become reassortant rotaviruses major two genes in these stocks.
Table 2: Genomic constellations of G6P[9] rotavirus (KF17) and prototype strains. View Table 2
Rotavirus bovine type G6 geno type detected in this study was generally rare. G6P[9] was originally isolated from an Italian child with diarrhea [5] and has subsequently been reported in the United States [6], Hungary [7], Japan [8], Australia [14,15], and Tunisia [16]. The authors [8] considered that their isolates represented reassortment events between bovine-like human rotaviruses and human/feline AU-1-like rotaviruses. G6 is a common genotype in cattle/buffalo [17], sheep [18], and goats [19,20] and has been identified sporadically or at a low prevalence in rabbits and pigs [21].
G6P[14] in Egypt [22] and G6P[9] in Burkina Faso [23] in either was reported as a rare virus. Case 24 in this study without previous contact with obvious bovine did not identify the source of infection. In the phylogenetic analysis the KF17 VP7 gene was located in a human lineage including other human G6 strains. Similarly, all other KF17 genes except for the NSP3 gene were relatively closely related to at least one of the human G6 RVs reported in Europe and the U.S. In a study in Miyagi Prefecture G6P[9] (M72S11) per sample was found from a 2-year-old toddler in 2011 (Figure 2 and Figure 3) [24]. Also G6P[9] have been detected from more than 3 cats of Mie Prefecture. These findings suggest that human G6 RV which had occurred by reassortment between human and bovine RV are distributed worldwide, despite low prevalence. Since genotypes of a few gene segments are different among those human G6 strains, this suggests that G6 rotaviruses may occur independently in different locales or countries through reassortment among local strains.
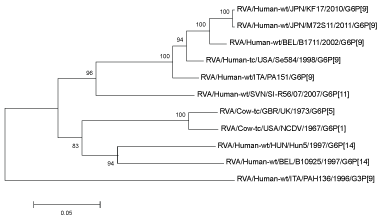 Figure 2: Phylogenetic analysis of VP7 genes of G6P[9] (M72S11). View Figure 2
Figure 2: Phylogenetic analysis of VP7 genes of G6P[9] (M72S11). View Figure 2
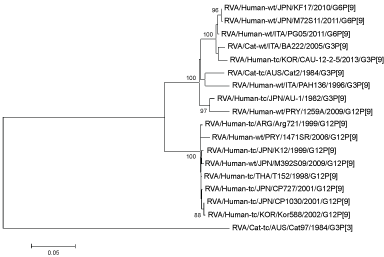 Figure 3: Phylogenetic analysis of VP4 genes of G6P[9] (M72S11). View Figure 3
Figure 3: Phylogenetic analysis of VP4 genes of G6P[9] (M72S11). View Figure 3
Although infections with Feline Rotaviruses (FRV) rarely cause severe illness in cats [25,26], FRVs have captured attention as perpetuating, albeit infrequent, sources of human disease. Human RV with genetic homology to FRV have been isolated from widespread geographical locations, including Japan [27,28], Israel [29], Tunisia, and the United States [30]. Additionally, putative human/feline reassortant rotaviruses have been identified in children in Italy [31].
Two G and P genotypes identified combinations carried by rotaviruses circulating in the cat population were G3P[9] and G6P[9]. Both G3P[9] and G6P[9] genotypes have been isolated from people. G3P[9] is a recognized feline genotype (AU-1-like and BA222-like genotype constellations [28,32]. Ferine RV (G6P[9] and G3P[9]) were detected at a low prevalence (3.0%) in the cat population in the United Kingdom. G6 was the more prevalent genotype (84%) and was detected in Scotland, the Midlands, and Cornwall, which are geographically distinct regions encompassing the length of the United Kingdom. This is similar to estimates from other countries including Japan [8] but showed regional and seasonal variations. Longitudinal sampling strategies based on this prevalence could be used to investigate transmission dynamics in more detail. The absence of an association between RV infection and diarrhea in cats is in stark contrast to humans. Infection has also been associated with diarrhea and decreased productivity in cows [33,34], suckling pigs, and horses [35]. Asymptomatic infections are reported, although their importance in transmission is not well understood due to the lack of population-based studies [36]. In cows, asymptomatic individuals shed similar viral titers to those of clinically infected individuals, and the roles of virulent and avirulent strains of RV have been postulated [37,38]. Asymptomatic infection may be a reflection of the nature of the relationship of G6P[9] and G3P[9] with the feline host [39].
The possibility of G6 feline origin at some historical point has been proposed. With clustering of published human G6 genotypes with our feline G6 genotypes, rather than with published bovine G6 genotypes [40], it was strongly suggests that G6P[9] genotypes were examples of zoonotic or anthropozoonotic transmission between cats and people. Whole-genome sequencing to further explore the relationship between the G3P[9] and G6P[9] genotypes identified in human and animal rotaviruses is under way.
G6P[9] is a relatively common feline rotavirus exists at low prevalence in cat population in Japan. Diarrhea and age are not risk factors for infection. Transmission events between cats and people likely exist, although they are infrequent and do not cause outbreaks of disease. The surveillance of RV in domestic pet population is important for investigating rotavirus genetic diversity, elucidating the role of asymptomatic carriage, exploring zoonotic risk, and monitoring the potential role of nonhuman rotaviruses in the evolution of rotavirus.
A very rare G6P[9] type related to bovine rotaviruses was also detected. It was strongly suggested that G6P[9] genotypes are examples of zoonotic or anthropo-zoonotic transmission between cats and people. Further epidemiological studies are necessary concerning this strain. Investigation of the epidemic strain of rotavirus after introduction of a vaccine is also considered necessary.
The authors express their gratitude to Nobumichi Kobayashi at the Department of Hygiene, Sapporo Medical University School of Medicine, Japan, for their valuable technical assistance and comments.
No commercial relationship or potential conflict of interest related to the submission of this manuscript.