Toxoplasma gondii genotypes display high genetic diversity in South America with levels of diversity not yet seen in Europe. Indeed, highly virulent strains even for immunocompetent humans have been described. However, limited or no information appears to be available dealing with strains of T. gondii circulating in Uruguay. In order to understand the Toxoplasma population in Uruguay, 252 samples of sera from seropositive individuals living in Uruguay were selected for serotyping analyses. An Enzyme-Linked Immunosorbent Assay (ELISA) based on the antibody response to specific peptides derived from T. gondii strains was used. Our findings predicted the presence of 34 discrete serotype profiles. Classical archetypes, serotype II and serotype III, were found for 8.3% and 2.4% respectively. Intriguingly, 29 different atypical serotype profiles were predicted, and non-reactive serotype profile was found in 12.3% of the samples. There appears to be a high genetic diversity within Toxoplasma population in Uruguay, similar to that known for neighboring countries in South America.
Toxoplasma gondii, Serotyping, Serotype prevalence
Toxoplasmosis presents the highest human incidence amongst the parasitic zoonoses [1]. Toxoplasma gondii is a parasitic protozoa with huge veterinary and medical importance, and a widespread distribution in a variety of livestock, wild animals and humans. Genetic diversity of T. gondii has been extensively studied over the last years. First studies suggested a clonal population structure comprising three major lineages (I, II and III) in Europe and North America [2]. Nevertheless, recent studies using multilocus typing show that the parasite exhibit a more complex genetic structure [3-5], with the highest diversity in South America. In Brazil, namely, parasite genetic structure includes unique genotypes [6-8], and several new clonal lineages [6]. Some atypical genotypes, notably those circulating in the Amazonian rain forest are implicated in severe cases of infection and eventually death in immunocompetent patients [9-12]. Other atypical genotypes have also been associated with cases of ocular toxoplasmosis, particularly in South Brazil and Colombia [13,14]. Notably, in the same countries, severe cases of congenital toxoplasmosis are more frequent than in Europe, likely due to different genotypes [15].
In Uruguay, no information exists regarding strains of T. gondii circulating in humans. Uruguay is a country located in the southeast of the South American continent sharing borders with Brazil in the north and northeast, and Argentina in the west, with 88% of its population of European descent. The economy is largely based on agriculture, the state sector and on trade, particularly in agricultural exports. To understand the Toxoplasma genetic diversity in Uruguay, human serum samples were serotyped using an Enzyme-Linked Immunosorbent Assay (ELISA) based on the antibody response to specific peptides derived from T. gondii strains [16-19]. Herein, we describe Toxoplasma serotype profiles in a survey in Uruguay, and analyse the data according to the geographical and socio-economical context.
Serum samples collected in the scope of the toxoplasmosis screening program were sent to the Parasitology Unit of the Public Health laboratory of Uruguay for serological confirmation. Two hundred and fifty-two human sera known to be seropositive for Toxoplasma were included in the present study. Of these 252 sera, 242 were provided during pregnancy; the other 10 were from asymptomatic or lightly symptomatic cases of toxoplasmosis. Genotypes of the strains were not known. The geographical origin of the cases providing the sera was: Department of Artigas (two samples), Rivera (one sample), Rio Negro (23), Tacuarembó (15), Cerro Largo (20), Durazno (5), Flores (4), Treinta y tres (7), Colonia (39), Canelones (22), Lavalleja (3), Maldonado (3), Rocha (2), Florida (4), San José (1) and Montevideo (73 suburban; 28 urban region) (Figure 1). Twelve sera from people negative for Toxoplasma were used to establish cut off values.
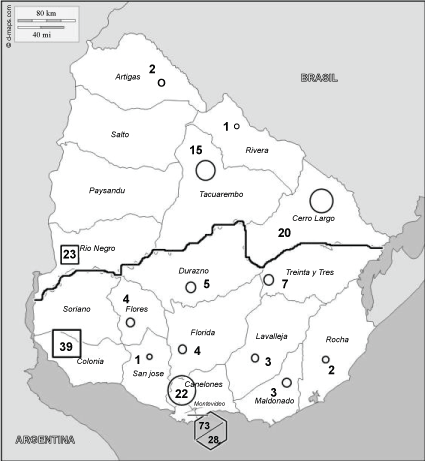 Figure 1: Map of Uruguay. Regions from which serum samples were collected and number of serum samples by department are marked as well as number of serum samples from Montevideo suburban (73) and urban (28) region. North and South region are divided by the black line □ represents agricultural regions; ○ represents animal husbandry regions,represents Montevideo.
View Figure 1
Figure 1: Map of Uruguay. Regions from which serum samples were collected and number of serum samples by department are marked as well as number of serum samples from Montevideo suburban (73) and urban (28) region. North and South region are divided by the black line □ represents agricultural regions; ○ represents animal husbandry regions,represents Montevideo.
View Figure 1
Seven strain-specific synthetic peptides derived from GRA6 and GRA7 loci as well as a peptide control were used, as already described [18,19] (Table 1). Four peptides (GRA6II, GRA6I/III, GRA7I, GRA7III) had polymorphisms specific for Toxoplasma archetypal lineages I, II and III. Other three peptides were selected according to known South American and African strain sequences, i.e. Am6, Af6 and Am7.
Table 1: Amino acid sequences of synthetic peptides derived from GRA6 and GRA7 loci. View Table 1
For each peptide, ELISAs were performed as previously described [18,19]. Briefly, immobilizer amino plates (Nunc, Denmark) were coated with each peptide diluted to 10 g/ml in 0.05 M carbonate/bicarbonate buffer pH 9.6 by incubation overnight at 4 ℃. Wells were blocked with PBS/BSA 3%. Sera were diluted to 1/50 in PBS/Tween 0.3%/BSA 3% and anti-human IgG alkaline phosphatase conjugate (Pierce, USA) was diluted to 1/5000 in PBS/BSA 3%/Tween 0.3%. Reaction was developed with p-nitrophenyl phosphate as the substrate. Absorbance at 415 nm was measured. Optical Density (OD) index was calculated by subtracting the OD of the peptide control from the OD of each peptide. Cut off was set on the mean absorbance readings of negative sera plus two standard deviations (2SD). Sera were considered positive for these peptides when the OD index was equal or higher than established cut-off (0.061, 0.014, 0.496, 0.277, 0.119, 0.769 and 0.101, respectively).
To understand Toxoplasma strains circulating in Uruguay, we analysed separately urban and suburban areas of Montevideo, North and South regions, and agricultural vs. husbandry regions. The classification of agricultural regions was based on Regiones agropecuarias del Uruguay (2015), Ministerio de Ganaderia Agricultura y Pesca del Uruguay (http://www.mgap.gub.uy/dieaanterior/regiones/Regiones2015.pdf). Geographical regions were defined based on the ecoregions defined by Brazeiro, et al. [20] and by the Rio Negro. The west region of the sedimentary basin of the Uruguay river is mainly agricultural. Animal husbandry across the country is not homogeneous. South of the Rio Negro (southern region), beef cattle husbandry coexists with other agricultural activities, mainly milk production and horticulture. The northern region is mainly a cattle and sheep husbandry region (Figure 1). Statistical analysis was performed using Graph Pad Prism 6 software. Fisher's exact test was used to compare atypical with non-atypical serotypes and Non-Reactive (NR) to all other serotypes (reactive serotypes). Serum samples from Montevideo were analysed separately. The remaining departments of the country were divided according to the main economic activity (animal husbandry vs. agriculture) and the geographic origin (Northern vs. Southern region). The following conditions were evaluated:
a) Atypical vs. non-atypical serotypes and association with economic activity
b) Atypical vs. non-atypical serotypes and association with geographic origin
c) Atypical vs. non-atypical serotypes for agricultural regions and association with geographic origin
d) Atypical vs. non-atypical serotypes for animal husbandry regions and association with geographic origin
e) Atypical vs. non-atypical serotypes for Montevideo urban and suburban districts
f) Reactive (atypical and non-atypical) vs. non-reactive serotypes and association with economic activity
g) Reactive vs. non-reactive serotypes and association with geographic origin
h) Reactive vs. non-reactive serotypes for agricultural regions and association with geographic origin
i) Reactive vs. non-reactive serotypes for anmal husbandry regions and association with geographic origin
j) Reactive vs. non-reactive serotypes for Montevideo urban and suburban districts
A P value < 0.05 was considered statistically significant.
Thirty-six different serotype profiles were identified among the 252 sera from Uruguay (Table 2). No atypical profiles, compatible with clonal lineages type I, II, and III, were found in 13.9% of samples (35/252). Serotype II was found for 21 patients (8.3%). This serotype was defined by the exclusive reactivity with the peptide GRA6II. Serotype III was found for five samples (2%), with two different reactive profiles. Three out of these five samples reacted exclusively with the peptide GRA7III, and the other two reacted with peptide GRA7III as well as GRA6I/III. A GRA6I/III profile (exclusive reaction with peptide GRA6I/III) was also found for seven patients (2.8%), which suggest the existence of an allele I or III for GRA6 locus.
Table 2: Serotype profiles found among 252 human serum samples from Uruguay View Table 2
Two other profiles were found for two samples: II/I/III and II/III (1 each). Profile II/I/III was defined by the reaction with peptides GRA6II and GRA6I/III. Such reactivity may suggest the existence of a mixed infection, with a type II and a type I or III strain or of an infection with a recombinant strain. Profile II/III was defined by the reaction with peptides GRA6II and GRA7III. Twenty-nine different conjugations between the peptides specific for the clonal lineages and the nonarchetypal ones were found. These serotypes were defined as atypical (atypical 1 to 29). Altogether, these atypical serotypes were found in 73.8% of samples (186/252). The two most prevalent atypical serotypes were: atypical 6 and atypical 19. Serotype atypical 6, seen with 40 sera (15.9%) was defined by the reaction of peptides GRA6II, GRA6I/III, Am6 and Af6. Atypical serotype 19, found for 38 sera (15.1%) was defined by the reaction of peptides GRA6I/III, Am6 and Af6 (Table 2). Four other atypical serotypes were found for more than 10 seropositive cases of the 252 sampled. Atypical 5 serotype was found for 21 (8.3%), atypical 1 was found for 19 (7.6%), atypical 4 was found for 17 (6.7%) and atypical 2 was found for 10 (4%). The remaining atypical serotypes were found for less than 10 serum samples (Table 2). Another serotype was also found for 31 sera (12.3%), defined as Non-Reactive serotype (NR). Sera from persons with this serotype did not recognize any peptide.
The sera were grouped into two categories according to the predominant economic activity of the region: 1) Agriculture and 2) Animal husbandry (Figure 1). Among 89 sera, from the animal husbandry regions, 14.6% (13 patients) displayed non-atypical serotypes, and 75.3% (67 patients) had atypical serotypes (Table 3). For agricultural regions, non-atypical serotypes were also found for 9 cases (14.5%), while atypical serotypes were found for 74.2% (46 patients). These differences were not statistically significant (Table 3). Furthermore, no significant statistically difference was found when economic activity was associated with geographic origin (Table 4). Some profiles were exclusively found for animal husbandry regions: serotypes II/III, II/I/III, atypical 8, 9, 10, 17, 26 and 28. Atypical serotype 29 was exclusively found for agricultural region. NR serotype was found for 9 serum samples (10.1%) from animal husbandry region and for 7 samples (11.3%) from agricultural region.
Table 3: T.gondii serotypes and association with economic activity and geographic origin. View Table 3
Table 4: T. gondii serotypes for agricultural and animal husbandry regions and association with geographic origin. View Table 4
Serotyping results were also analysed according to the geographical origin of the patients (Figure 1). Among 61 serum samples from North region, 62.3% (38 patients) had atypical serotypes and 23% (14 patients) had non-atypical serotypes (Table 3). For the Southern region, atypical serotypes were found for 82.2% (74 patients) and non-atypical serotypes were found for 10% (9 patients) (Table 3). The differences were statistically significant. However, when the Northern and Southern regions were separately associated with the economic activity, statistically significant differences were not apparent| (not shown). Some profiles were exclusively found for the Northern region: serotypes atypical 8, 9, 10, 28. Serotypes atypical 3, 17, 26 and 29 were exclusively found in the South. NR serotype was found for 9 serum samples (14.8%) from the Northern region and for 7 samples (7.8%) from the South.
Sera from cases from Montevideo were analysed separately; the urban and suburban regions were compared (Table 5). Non-atypical serotypes were present in both groups. However, only two out of 28 samples (7.1%) from Montevideo urban region had non-atypical serotypes (II and III). Atypical serotypes were found for 75% of urban serum samples (21 cases) and for 72.6% of suburban samples (53 patients). Serotype atypical 23 was exclusively found in the urban region of Montevideo. Serotypes atypical 11, 12, 15, 16, 20, 22, 24 and 27 were exclusively found for the Montevideo suburban area. NR serotypes were found for 5 samples (17.9%) from urban region and for 10 samples (13.7%) from the suburban areas.
Table 5: T. gondii serotype in Montevideo and association with urban districts. View Table 5
Serotyping has been pointed as a promising tool for classifying Toxoplasma gondii genetic structure. Serotyping has the advantage of not requiring isolation of parasite or its DNA, since it is based on the antibody recognition of strain-specific polymorphic peptides [16-19], and has also the potential to be used as a screening tool for typing in areas where archetypal T. gondii infections prevail [21]. In this work, serotyping was used with the objective of characterize the genotypes of Toxoplasma circulating in Uruguay; to our knowledge, no information has previously been reported on this topic. Genotyping studies of Toxoplasma isolates from South American countries have shown that for that geographical region genotypes different from the clonal type II and III lineages are mainly found [3-5]. More recently, type II was found in Brazil [22,23] and Argentina [24-26], although in markedly lower frequency than in Europe. Using serotyping methods, previous reports also described profiles coincident with the archetypal lineages I and III in South America [17,27]. In Uruguay, serotype profiles compatible with the archetypal lineages II and III were found in 13.5% of the seropositive samples. For 2 other patients, a mixed infection with typical strains could be suggested. Unexpectedly, prevalence of these non-atypical serotypes was higher in the north than the south of Uruguay north borders with Brazil and Argentina may explain this difference. Another explanation may be that the north region is essentially an animal husbandry region with smaller biodiversity [28], which could contribute to a higher success of typical strains well adapted to livestock. In Europe, type II strains account the majority of congenital cases during pregnancy [29,30], and ocular toxoplasmosis cases in France [31] and Germany [32]. Uruguay has an important trade exchanges with Europe. When ships from Europe arrived in Montevideo port they could introduce T. gondii in the area through cargo infested with oocysts as well as infected mice and cats [33]. Our hypothesis that more typical strains would be found in the urban area of Montevideo was not confirmed: Only 7% of the 28 patients from this area close to the port presented a non-atypical serotype profile. The proportion is higher in more distant rural regions. The T. gondii strains introduced from Europe by the maritime trade over the last centuries could have been perpetuated in Uruguay by the extensive animal husbandry, since Uruguay has an important wool and meat processing industry. Introduction of non-atypical strains in Uruguay follows the livestock introduction in the country by the 17th century. However, the life cycle of the native atypical strains was perpetuated due to the country rich biodiversity, especially in the South. However, our results do not suggest that animal husbandry alone contributes to the selection and dissemination of certain types of strains restringing others, since differences associated with economic activity were not statistically significant.
However, there are limitation with our serotyping; serotype II was defined by the recognition of a single peptide from only one Genetic Marker (GRA6), and serotyping, based on a single marker, can lead to misclassification. Also, infections due to nonarchetypal strains can be misclassified as type II strains or type I or III strains since GRA6 C-terminal peptides specific for type II and type I or III strains cross-react with serum samples from patients with infections caused by nonarchetypal strains [18]. Consequently, sera classified as non-atypical serotypes may indeed be atypical serotypes and a possible serotype misclassification can also justify the higher prevalence of non-atypical serotypes in the north as well as in the suburban districts of Montevideo. To confirm that samples positive for GRA6II peptide are actually serotype II, other peptides from different antigens, with polymorphisms specific for type II strains, must be explored. A high percentage of seropositive cases, where sera reacted with the peptides specific for the atypical strains also reacted with one or more archetypal-specific peptide. Cross-reactivity between peptides specific for the archetypal lineages and atypical strains has been reported previously [18]. A serotyping study using serum samples from 18 human infections due to nonarchetypal strains showed that those strains were misclassified as infection with archetypal strains [18]. Serotyping of feline sera showed that cats infected with atypical strains reacted with peptides specific for type I and type III [21]. Similarly, peptides specific for the atypical strains also recognize archetypal strains [19]. Sequencing of GRA6 locus from 19 discrete haplotypes and of GRA7 locus from 14 haplotypes showed that GRA6 and GRA7 C-terminal region shared polymorphisms with the archetypal genotypes [19]. This could explain the cross-reactions observed. By contrast, a limitation of classical genotyping methods is that in the case of mixed infections, in a particular host, it may favor the isolation of one strain, restringing others. Serotyping, by recognizing strain-specific peptides, allows predictions for mixed infections. Different combinations of reactivity with peptides specific for different strains suggest the possibility of mixed infections [14]. Among the sera studied here, only two samples reacted with different peptides specific for the archetypal lineages, suggesting the existence of a mixed infection.
Non-reactive serotype was described for the first time for German patients with ocular toxoplasmosis [32]. Sera from cases positive for T. gondii that did not reacted with any serotyping peptides were also observed in our present study. The percentage of NR serotype was 12.3% whereas among German ocular toxoplasmosis patients the percentage was 44%, compared with 7% for non-ocular toxoplasmosis. The use of peptides specific for atypical strains does not explain this difference, since if those peptides were not used, the percentage of NR serotype would rise only to 16%. It would be of interest to analyse which genotype is associated with this NR serotype in Uruguay, in order to understand if there is a geographical difference between NR serotype in Uruguay and NR serotype found for German patients.
The results from this study suggest that Toxoplasma population in Uruguay has a high genetic diversity predicted by the different serotype profiles found. The high genetic diversity found for Toxoplasma strains in South America has been explained by the high biodiversity found for tropical rain forest [33]. Uruguay is not embraced by the tropical rain forest. However, its sole land border is with Brazil to the north and northeast; a highly diverse Toxoplasma population occurs in Brazil. Our results suggest the migration and expansion of nonarchetypal Toxoplasma strains to other countries of South America such as Uruguay, and emphasize the need for active monitoring of prophylactic vigilance during pregnancy. In conclusion, similarly to the genotyping, serotyping must be based in the use of multilocus peptides specific for each serotype. Serotyping has the potential to predict the profile of T. gondii strain circulating in the human residents, including establishing the presence of mixed infections or infections by nonarchetypal strains.
The authors would like to thanks Fundação para a Ciência e a Tecnologia for UID/MULTI/00211/2013. SS has a post-doctoral fellowship SFRH/BPD/103788/2014 from Fundação para a Ciência e a Tecnologia, Portugal.
This study was conducted on a group of anonymous human serum samples.