To investigate Twist1 expression and biological role in a series of diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL). Twist1 has been shown to promote oncogenesis in solid tumour.
Twist1 expression was investigated by both real-time quantitative polymerase chain reaction (RT-QPCR) and immunohistochemistry in 8 samples from reactive lymphoid tissues, 38 samples from DLBCL and 25 samples from FL patients. Results were correlated with clinicopathologic features. Both RT-QPCR and immunohistochemistry demonstrated increased Twist1 in DLBCL and FL in comparison to benign lymph nodes, where only stromal cells expressed Twist1. Twist1 was higher in FL. In DLBCL there was a tendency towards a higher level in the non-germinal-center subtypes. Twist1 expression did not correlate with an adverse prognosis and any clinicopathologic features.
Twist1 is overexpressed in lymphomas and immunohistochemistry is a valuable tool in routine practice to asses Twist1 expression. Twist1 expression in stromal component suggests a role of microenvironment in its up-regulation and in B cells transformation. Although Twist1 expression did not correlate with an adverse prognosis it interacts with other pathways involved in prognosis and drug resistance in lymphomas. Further investigations are needed to target it in future experimental trials.
Twist1, Follicular lymphoma, Diffuse large B-cell lymphoma, Oncogenesis
Twist1 is a transcriptional factor member of the basic helix-loop-helix (bHLH) class B family that has been shown to be important in embryogenesis. Specifically, Twist1 regulates mesodermal development [1]. Mutations in Twist1 cause Saethre-Chotzen syndrome, characterized by craniofacial and limb anomalies [2,3]. Expression of Twist1 is regulates by diverse signaling pathways especially linked to inflammatory-related cytokines via NFκB pathways [4] and to stress conditions via STAT3 pathways [5]. Once expressed Twist1 might act directly as a transcription factor, might activate gene expression through interaction with other transcription factors or might inhibit transcription through sequestration of transcription factor or by epigenetic modulation [6]. In solid tumors, where Twist1 may be overexpressed, it induces epitheliomesenchymal transition (EMT), promotes angiogenesis and metastatic potential [7-10]. A higher expression level of Twist1 in numerous solid tumors correlates with early dissemination of the disease and a poor clinical outcome [11-13]. In breast carcinoma Twist1 has been shown to deregulate P53 response to γ-radiation, to induce angiogenesis and to correlate with chromosomal abnormalities [14,15]. Twist1 is also able to neutralize senescence programs through inhibition of p53 [16], and to block apoptotic response through MYC interaction [17], providing cells with a growth advantage and rendering them resistant to chemotherapeutic agents, such as taxol and vincristine [16,18]. Despite the role of Twist1 in oncogenesis of solid tumor, the expression and the biological significance of TwistT1 in hematopoietic cancer have not been extensively studied. In hematopoietic system, the expression of Twist1 is largely observed in CD34+ hematopoietic stem cells [19,20]. Twist1 is also expressed in follicular dendritic cells which are specialized mesenchymal-derived stromal cells found in the germinal centers of lymphoid tissues that trap and present immune complexes at their surface to B cells, enabling their maturation [21]. When looking at T cells, Twist1 expression is upregulated and persistent in CD4+ T cells from inflamed tissues of patients with chronic inflammation (Crohnγs disease, ulcerative colitis, rheumatic disease) [22]. The expression of Twist1 in B lymphocytes has been poorly described. In T cell lymphoma, increased Twist1 protein expression is observed in advanced mycosis fungoides (MF) and sézary syndrome (SS) [23,24]. Twist1 is also aberrantly expressed in ALK-positive anaplastic large cell lymphoma (ALK + ALCL) and contribute to this lymphoma invasiveness and decreased sensitivity to Crizotinib [25]. Twist1 expression in B cells lymphoma has been poorly investigated. However, a study showed that Twist1 expression is increased in diffuse large B-Cell lymphoma (DLBCL) [26]. Diffuse large B-cell lymphomas (DLBCL) represent a heterogeneous group of diseases with variable clinical, pathological and genetic features. DLBCL patients may be stratified into different risk-groups according to clinical parameters and the international prognostic index (IPI) remains actually the best tool to predict response to treatment but does not reflect the molecular heterogeneity of the disease within each IPI subgroup [27,28]. A number of biological factors have been identified to have a prognostic significance in DLBCL. DLBCL with a GC phenotype and pattern of gene expression have a more favourable outcome than non-GC subtype [29]. Similar, expression of BCL2 and P53 protein or the presence of the P53 mutation have been shown to have an adverse effect in DLBCL [30-34]. However, discrepant results were published. Therefore, new prognostic biomarkers need to be defined based on the analysis of the molecular heterogeneity of DLBCL. To our knowledge, role of Twist1 has been poorly investigated in hematologic malignancies. Preliminary data obtained from sample of patients with B cell lymphoma derived from Origene (Lymphoma cDNA Array II LYRT102, Origene Technologies, Inc., Rochville, MD, USA) [6] demonstrated that Twist1 was overexpressed in several type of B cell lymphoma, especially in DLBCL and follicular lymphoma (FL).
Based on these data, in this study, we quantified Twist1 in DLBCL and FL by real-time quantitative polymerase chain reaction (QT-PCR) on frozen sample and assessed Twist1 protein expression with immunohistochemistry on fixed formalin paraffin embedded (FFPE) tissue. Our aim was first to investigate the frequency of the expression of Twist1 in these lymphomas. Second to correlate results of QT-PCR with those of immunohistochemistry that allowed to study, in situ, Twist1 protein expression with distinction between tumoral and micro-environment cells component. Third, to investigate potential correlations of Twist1 expression with biological parameters of these lymphomas.
The study population consisted of 63 lymphoma patients and 8 patients with reactive lymphoid hyperplasia (benign lymph node hyperplasia). The 63 lymphoma samples were from patients with DLBCL (38/63) or FL (25/63) from whom both FFPE and frozen tissues were available. All of the patients had histologically confirmed DLBCL or FL. All FL were grade 1/2 FL according to the WHO Classification [35]. DLBCL were classified according to the cell of origin (COO) based on immunohistochemical Hans algorithm that defined center B-cell (GC) DLBCL and non-GC DLBCL. The sample had been collected between 1996 and 2013 at the time of diagnosis. Diagnoses were reviewed by 2 experienced haematopathologists (VS, VCM). Detailed clinical information was collected for every patient, including staging at diagnosis defined by Ann Arbor, type of therapy, and follow-up with special attention to recurrence and status at last consultation. All patient with DLBCL received R-CHOP like chemotherapy whereas patients with FL received either R-CHOP like chemotherapy, Rituximab alone or were on therapeutic abstention. All clinical researches were conducted with written informed consent.
B lymphoma cell lines, BJAB, HBL1, SUDHL4, SUDHL6 and U2932 and breast carcinoma cell lines T47D were used as control. BJAB, SUDHL4 and SUDHL6 are GC cell lines. U2932 and HBL-1 are non-GC DLBCL-derived cell lines generously obtained from L. Staudt. Cell lines were cultured in RPMI 1640 medium (Life technologies) with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. T47D is a human breast tumor derived cell line graciously given by A. Puisieux cultures in maintained in Dulbecco's modified Eagle's medium (DMEM, life technologies), supplemented with 10% FBS, 1% penicillin and streptomycin, 1% Sodium Pyruvate (life technologies), 1% non-essential amino acids (life technologies), 1.44 ml Insulin at 37 ℃, 5% CO2.
Cells were lysed using 10 mM EDTA-radioimmunoprecipitation assay buffer containing protease inhibitors (Roche) and lysates were separated on 12% SDS-polyacrylamide gels (Bio-Rad) as previously described [36]. Mouse monoclonal antibody to Twist1 (Twist2C1a) was purchased from Abcam, stored at -20 ℃ and used at 1:750 dilution. Mouse moncoclonal antibody to β-actin (1:5000) was purchased from Sigma. Horseradish peroxidase-labeled goat anti-mouse IgG was purchased from Cell Signaling Technology. Signal was detected using FEMTO Western Blotting Substrate (Pierce).
RNA extraction was performed from cell lines and frozen sample tissues from lymphomas and benign lymph node hyperplasia. Sample were lysed according to the manufacturer's specifications (KIT SV total R NA Isolation System, Promega 50 preps #Z3105). cDNA synthesis was performed on 1 µg of total RNA after treatment with DNase (Promega, Madison, wt&) using Superscript II reverse transcriptse (Invitrogen, Breda, THE Netherlands) in a final volume of 24 µl. RT Q-PCR was performed using SYBER Green PCR Master Mix in an ABI-prism 7700 sequence detection system (Applied Biosystem, Nieuwerkerk aan den Ijssel, the Netherlands). The primer sequences (Invitrogen) for TWIST1 were forward 5-GCAGGACGTGTCCAGCTC-3 and reverse 5- CTGGCTCTTCCTCGCTGTT-3. The cycle parameters for this transcript and for the housekeeping gene GUS used for normalization were as follows: Denaturing for 15 s at 95 ℃; annealing and extension for 40 s at 60 ℃, for 40 cycle. Specificity of the PCR product was confirmed by agarose gel electrophoresis. The crossover point (Ct) values, defined as the cycle number at which each curve intersects the threshold detection value, were used to calculate gene-specific input mRNA amount of each tissue according to the calibration curve method. Data were evaluated using the SDS software version 1.9.1 (Applied Biosystems) and the second derivative maximum algorithm. Relative expression was calculated as 2(-(CTmean Twist1-CTmean Gus).
The tissue sample and cell pellet obtained from tumoral cell lines had been fixed in formalin and then embedded in paraffin. Representative tumor areas in the paraffin blocks were cut into 4-um sections. Immunostaining was performed using a Ventana Benchmark XT Autostainer (Ventana Tucson, AZ, USA). The following markers were used after appropriate antigen retrieval: CD20 (clone L26, Dako, Denmark A/S, 1:300), CD3 (clone F7.2.38, Dako, 1:20), CD5 (clone 4C7, Menarini, California USA, 1:100), CD10 (clone 56C6, menarini, 1:80), BCL6 (clone PG-B6P, Dako, 1:10), BCL2 (clone 124, Dako, 1:80), MUM1 (clone MUM 1p, Dako, 1:50), KI67 (clone MIB1, Dako, 1:100), Twist1 (Twist2C1a, Abcam, Cambridge, United Kingdom, 1:50).
The immunohistochemically stained slides were reviewed and analysed by 2 experienced haematopathologists (VS, VCM). Analysis was carried out using a light microscope. The immunoreactivity for TWIST1 was separately analysed in the stromal fibroblastic and tumoral lymphoid or reactive lymphoid cell compartments. Only nuclear positivity was considered significant. The immunreactivity for fusiform stromal cells was semiquantified as previously defined [37] as follows:
0-5% = Negative (0)
5-25% = Weak positivity (1+)
25-50% = Moderate positivity (2+)
50-75% = Strong positivity (3+)
> 75% = Very strong positivity (4+)
Finally, cases with score 0 were considered to be negative whereas cases with score 1+, 2+, 3+ and 4+ were considered to be positive.
For lymphoid reactive or tumoral cells, only the presence of nuclear negativity (0) or positivity (1) was assessed. Nuclear positivity was defined by percentage of staining > = 5%. Immunostaining was considered to be negative when percentage of staining was inferior to 5% [26].
Fisher's exact test, Kaplan-Meir method with the log-rank and Breslow and GraphPad software (Prism 6 for Mac OS X) were used to calculate the P value and to analyze survival data. Results were considered to be significant at P < 0.05.
Sixty frozen samples of both DLBCL and FL and 8 samples of benign reactive lymph nodes were available for these analyses. As shown in Figure 1A, Twist1 mRNA was undetectable in all except one reactive lymph node (0.0138 +/- 0.0087). The level of Twist1 mRNA was significantly higher in both DLBCL (0.0623 +/- 0.0113, p = 0.034) and FL (0.0815 +/- 0.0087, p = 0.0032) when compared to control. The highest levels of Twist1 mRNA were observed in FL compared to DLBCL, although the difference did not reach statistical significance (p = 0.2548). When DLBCL were stratified between GC type and non-GC type, there was a tendency towards a higher Twist1 mRNA level in the non-GC subgroup (0.0526 +/- 0.0146 versus 0.0695 +/- 0.0183, p = 0.237) (Figure 1B).
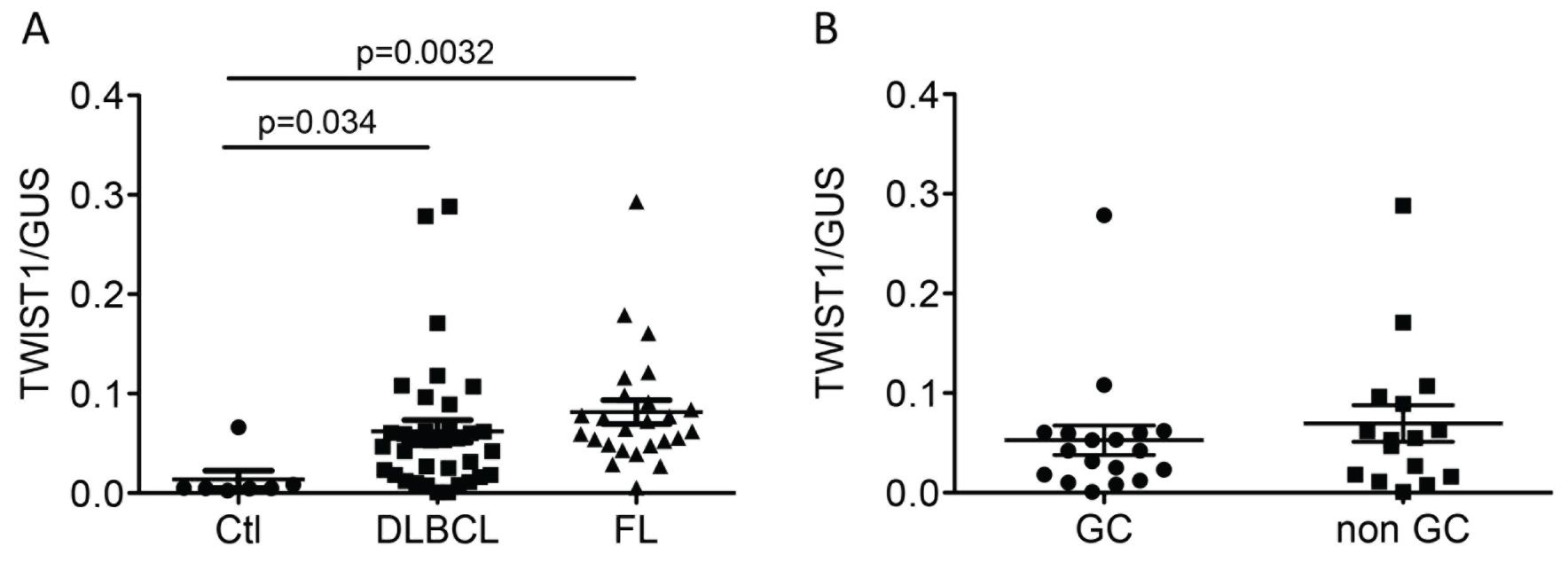 Figure 1: Twist1 mRNA levels in reactive lymph nodes used as control (Ctl), DLBCL and FL. Statistics were performed independently between groups and controls. A) Twist1 relative expression in Ctl, DLBCL and FL. Twist1 expression was significantly higher in FL and DLBCL; B) Twist1 was measured and compared in DLBCL GC (Germinal center) and non-GC subtypes. Twist1 expression was higher in the non-GC subtype, although it did not reach statistical significance. View Figure 1
Figure 1: Twist1 mRNA levels in reactive lymph nodes used as control (Ctl), DLBCL and FL. Statistics were performed independently between groups and controls. A) Twist1 relative expression in Ctl, DLBCL and FL. Twist1 expression was significantly higher in FL and DLBCL; B) Twist1 was measured and compared in DLBCL GC (Germinal center) and non-GC subtypes. Twist1 expression was higher in the non-GC subtype, although it did not reach statistical significance. View Figure 1
Twist1 protein expression was first verified, by WB, in breast tumor derived T47D cell line, further used as a positive control. Twist1 expression was repeatedly detected in non-GC DLBCL cell lines HBL-1 and U2932 but not in GC cell lines BJAB, SUDHL4 and SUDHL6 (Figure 2A). As shown in Figure 2B, Figure 2C, Figure 2D, Figure 2E, Figure 2F, Figure 2G and Figure 2H, Twist1 protein expression was found in the nuclei of T47D, HBL-1 and U2932 cell lines whereas BJAB was negative by immunohistochemistry.
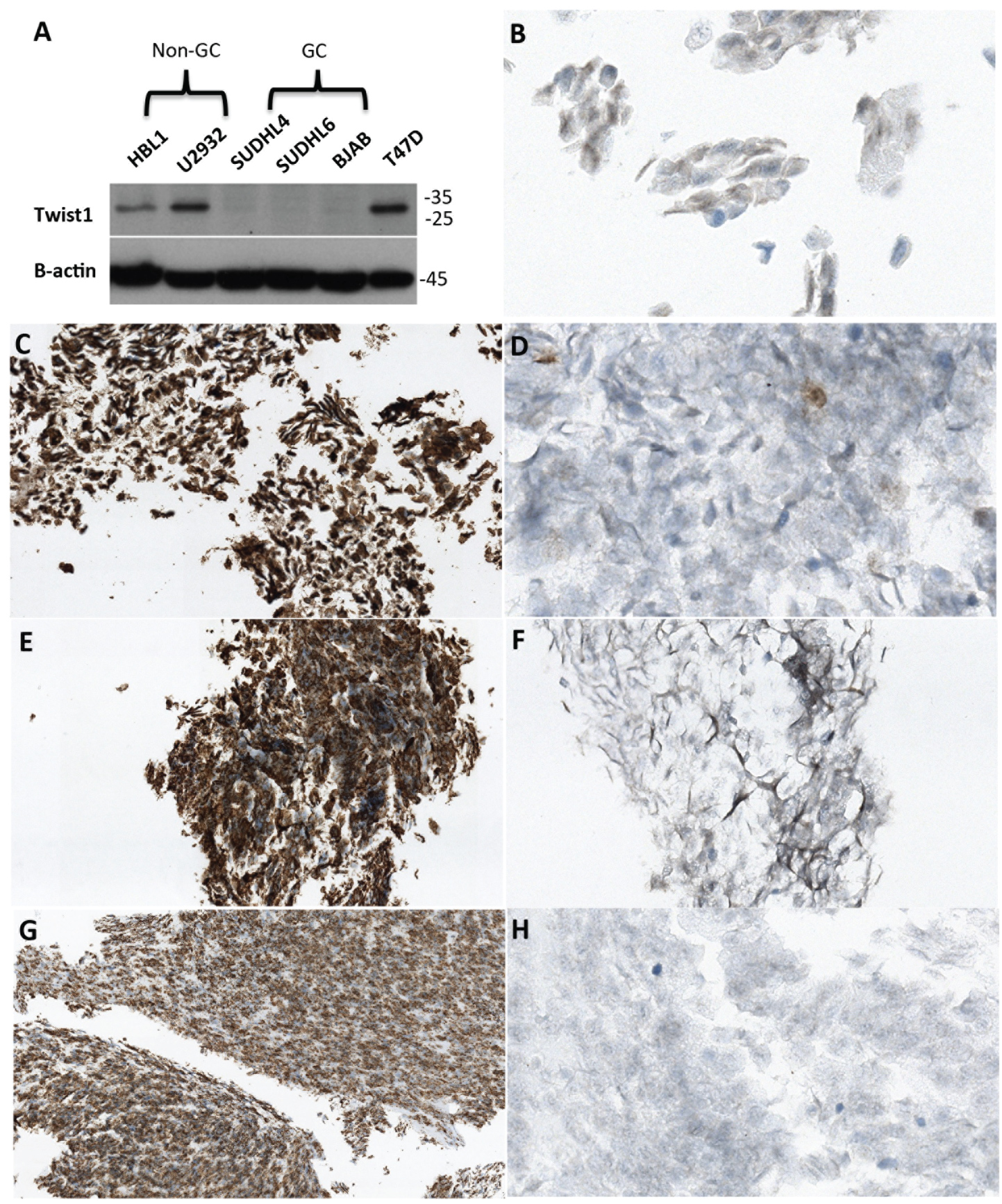 Figure 2: A) Twist1 expression was assessed by Western Blot in DLBCL derived cell line U2932 and HBL-1, SUDHL4, SUDHL6 and in B cell lymphoma cell line BJAB. Human breast tumor derived T47D was used as a positive control. Beta-actin was used as an internal control to verify the total amount of protein in the cell lysates. This Western Blot is representative of numerous experiments; B-H) Immunohistochemical studies showed that TWIST1 was found in T47D, HBL-1 and U2932. As expected BJAB was negative. The staining was nuclear. B) TWIST1 expression in T47D (X60); C) CD20 expression in U2932 (X20); D) TWIST1 expression in U2932 (X60); E) CD20 expression in HBL-1 (X20); F) TWIST1 expression in HBL-1 (X20); G) CD20 expression in BJAB (X20); H) TWIST1 expression in BJAB (X40). View Figure 2
Figure 2: A) Twist1 expression was assessed by Western Blot in DLBCL derived cell line U2932 and HBL-1, SUDHL4, SUDHL6 and in B cell lymphoma cell line BJAB. Human breast tumor derived T47D was used as a positive control. Beta-actin was used as an internal control to verify the total amount of protein in the cell lysates. This Western Blot is representative of numerous experiments; B-H) Immunohistochemical studies showed that TWIST1 was found in T47D, HBL-1 and U2932. As expected BJAB was negative. The staining was nuclear. B) TWIST1 expression in T47D (X60); C) CD20 expression in U2932 (X20); D) TWIST1 expression in U2932 (X60); E) CD20 expression in HBL-1 (X20); F) TWIST1 expression in HBL-1 (X20); G) CD20 expression in BJAB (X20); H) TWIST1 expression in BJAB (X40). View Figure 2
Immunohistochemistry was performed on FFPE patients' samples and FFPE cell pellets. For technical reasons (on-line supporting information), 7 out of the 8 included reactive benign lymph nodes and 28 out of the 38 included DLBCL were available and were stained for Twist1. All 25 FL were successfully investigated.
Twist1 protein expression was observed as a nuclear immunostaining in all positive cases (Figure 3). Twist1 was also detected in stromal cells from most DLBCL (17/28, 60.7%), FL (20/24, 83.3%) and reactive lymph nodes (4/7, 57.1%) (Table 1 and Figure 3). When comparing DLBCL subtypes (GC versus non-GC), stromal Twist1 expression tended to be lower in the GC subtype than in the non-GC subtype (p = 0.054) (Table 2).
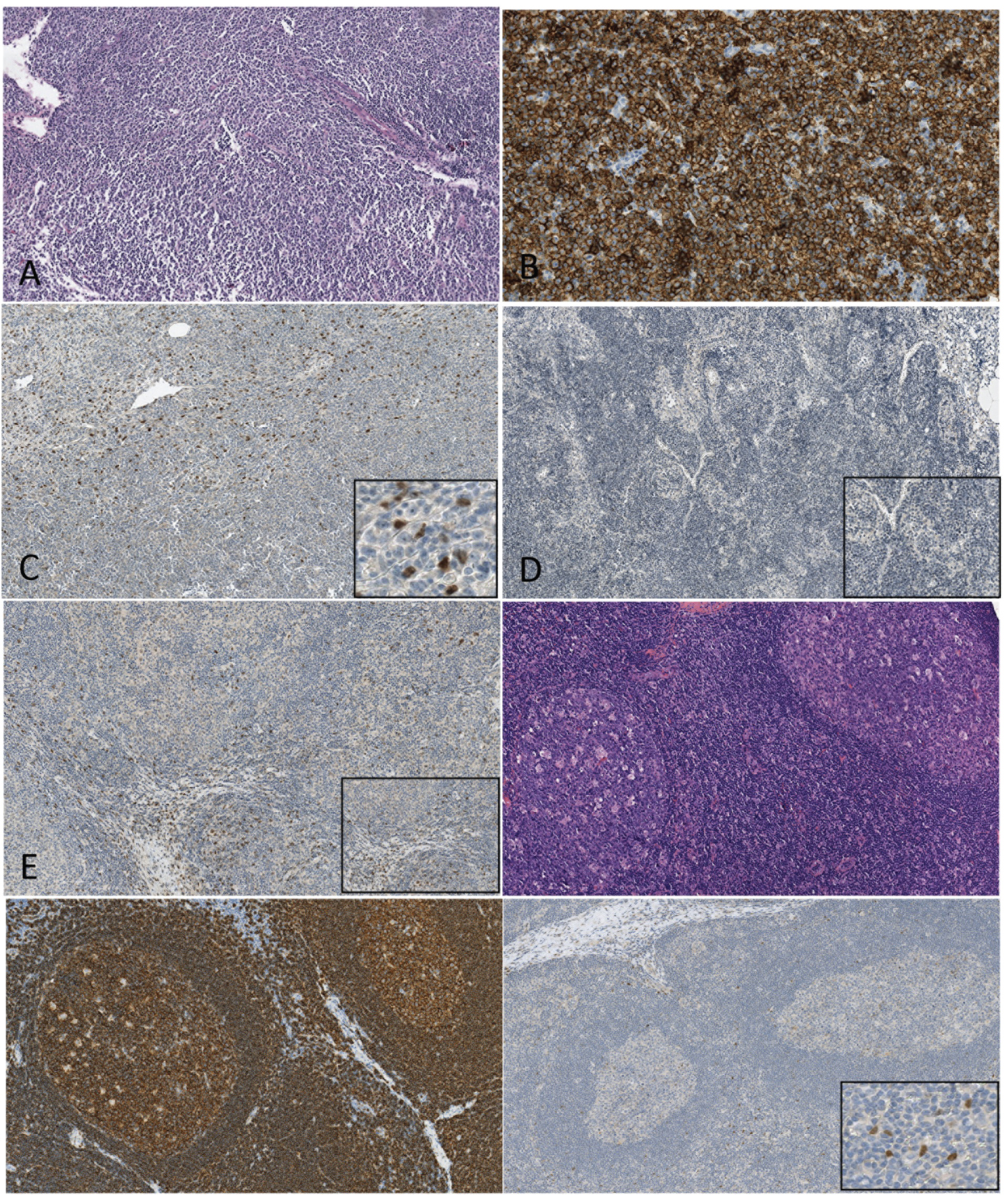 Figure 3: A, B, C Immunohistochemical staining of DLBCL. A) HEX10; B) CD20 X10; C) Twist1X10 and Twist1X20: nuclear expression of TWIST is found in nuclei of tumoral lymphoid and stromal cells. D, E) Immunohistochemical staining of FL. D TWIST X10 and TWIST X20: in this case, both tumor cells and fusiform cells are negative. F) TWIST X10 and TWIST X20: in this case, both tumor cells and fusiform stromal cells are positive. F, G, H - Figure E. Immunohistochemical staining of reactive begin lymph node; F) HEX10; G) CD20 X10; C) TWIST1X10 and H) TWIST1X20: nuclear expression of TWIST is found in nuclei of fusiform stromal cells while lymphocytes are negative. View Figure 3
Figure 3: A, B, C Immunohistochemical staining of DLBCL. A) HEX10; B) CD20 X10; C) Twist1X10 and Twist1X20: nuclear expression of TWIST is found in nuclei of tumoral lymphoid and stromal cells. D, E) Immunohistochemical staining of FL. D TWIST X10 and TWIST X20: in this case, both tumor cells and fusiform cells are negative. F) TWIST X10 and TWIST X20: in this case, both tumor cells and fusiform stromal cells are positive. F, G, H - Figure E. Immunohistochemical staining of reactive begin lymph node; F) HEX10; G) CD20 X10; C) TWIST1X10 and H) TWIST1X20: nuclear expression of TWIST is found in nuclei of fusiform stromal cells while lymphocytes are negative. View Figure 3
Table 1: Twist1 protein expression in B cells and microenvironment of DLBCL, FL and benign lymph node hyperplasia. View Table 1
Table 2: Twist1 protein expression in GC-type and non-GC type DLBCL. View Table 2
Nuclear Twist1 positivity in the lymphoid compartment was found in 83.3% (20/24) of FL, 35.7% (10/28) of DLBCL and in none of the reactive lymph nodes (0/7) (Table 1 and Figure 3). Twist1 expression in B cells was higher in tumoral B-cells component of FL than in reactive B-cells (reactive lymph nodes) (p = 0.0001, Table 1), and tended to be higher in tumor B-cells component of DLBCL than in reactive B-cells (although it the association wasn't statistically significant, p = 0.0835 Table 1). When comparing FL and DLBCL, Twist1 expression in neoplastic B cells was significantly increased in FL (p = 0.0007, data not shown). Moreover, Twist1 positive expression in neoplastic B cells was mostly observed in the non-GC subgroup (6/12, 50%) compared to the GC-subgroup (4/16, 25%), although the difference did not reach statistical significance (p = 0.2425; Table 3).
Table 3: Association between TWIST protein expression and clinico-pathologic factors in patients with DLBCL. View Table 3
The association between Twist1 expression and clinicopathologic variables in patients with DLBCL and FL are shown Table 3 and Table 4. The median follow-up for patients with DLBCL and FL was 10 months (range, 1-108 months) and 47 months (range, 1-144 months), respectively. Twist1 staining in tumor cells was used to stratify patients into 2 groups (positive Twist1 expression and negative Twist1 expression).
Table 4: Association between TWIST protein expression and clinico-pathologic factors in patients with FL. View Table 4
For patients with DLBCL, nuclear Twist1 positivity in neoplastic B-cells did not correlate with poorer overall survival (p = 0.7046 log rank; p = 0.7198 Breslow, data not shown) or poorer progression-free survival (p = 0.2855 log rank; p = 0.2805 Breslow, data not shown). Moreover, Twist1 protein expression did not correlate with any of the clinicopathologic variable tested (Table 3). Nevertheless, there was a trend towards an association between BCL2 protein expression and Twist1 protein expression in neoplastic B cells, since all the patients that did not expressed Twist1 protein did not expressed BCL2 protein either.
For patients with FL, nuclear Twist1 positivity in neoplastic B-cells did not correlate with poorer overall survival (p = 0.3028 log rank; p = 0.3994 Breslow, data not shown), although there was a trend towards poorer progression-free survival (p = 0.0734 log rank; p = 0.1207 Breslow, data not shown). Finally, Twist1 expression was not associated with any of the clinicopathologic variable tested (Table 4).
Twist1 is a transcription factor which overexpression in solid tumor correlate with EMT, promoted angiogenesis and metastatic potential. Twist1 is also able to neutralize senescence programs through inhibition of P53 and to block apoptotic response through MYC interaction providing cells with a growth advantage and rendering them resistant to chemotherapeutic agents, such as taxol and vincristine [16-18]. Little is known about Twist1 expression in B cell malignancies. With the aim to investigate a possible role of Twist1 in B-cell lymphoma, RT Q-PCR on samples derived from Origene (Lymphoma cDNA Array LYRT102, OriGene Technologies, Inc., Rochville, MD, USA) was previously performed in our team [6]. Overexpression of Twist1 was detected in DLBCL and FL samples, with higher level in FL samples compared to DLBCL samples [6]. Based on these preliminary data, we analysed expression and role of Twist1 in a series of DLBCL and FL with FFPE and frozen tissue available. We showed that mRNA level of Twist1 was higher in DLBCL and FL compared to reactive lymph nodes. In keep with our previous data, mRNA level of Twist1 was increased in FL compared to DLBCL [6]. We also showed a tendency to higher level in the non-GC DLBCL compared to the GC-DLBCL. Similar results were obtained with immunohistochemistry approach showing significant overexpression of Twist1 protein in both tumoral samples (with higher in FL than in DLBCL) compared to reactive lymph node. We also detected an increased expression of Twist1 protein in non-GC DLBCL subtypes compared to GC subtypes (although results were not statistically significant). So that immunohistochemistry with Twist1 antibody is a valuable tool in clinical practice to assess Twist1 expression in B cell lymphomas.
Twist1 overexpression is known to neutralize senescence through P53 inhibition and to block apoptosis through MYC pathway. Thus, Twist1 activation results to a survival advantage for lymphoid cells. P53 mutations occurred in about 7% of FL and 19% of DLBCL [33] and are associated with poor survival [32,33]. Approximately 5-10% of DLBCL harbour a 8q24/MYC rearrangement. DLBCL patients with MYC break have a significant inferior prognosis than DLBCL patients without MYC break [38,39]. It may be speculated that in some circumstances, MYC and P53 interact with Twist1 overexpression, resulting in malignant transformation by way of apoptosis inhibition. In Myelodysplastic syndrome (MDS) interaction between Twist1 and P53 has been demonstrated and is associated with resistance to apoptosis [19]. Some authors observed an association between increased Twist1, c-MYC and P53 expression in sézary syndrome (SS) and mycosis fongoïde (MF) [23] and have correlated these abnormal expressions with MF/SS clinical stages.
One hypothesis for overexpression of Twist1 in non-GC DLBCL compared to the GC-subtypes may be suggested. NFκB pathway is constitutively activated and STAT3 expression [40] is usually triggered in non-GC subtypes [41]. Interestingly these both pathways interact with Twist1. It has been shown that Twist1 overexpression is connected to cell resistance to chemotherapeutic agents, such as vincristine, and it is partly because of activation of NFκB pathway [6,42]. Interestingly non-GC DLBCL have poorer prognosis than GC subtype and poorer response to R-CHOP (Rituximab- Cyclophosphamide, Hydroxydaunorubicin, Oncovin (vincristine), Prednisone) including vincristine.
When comparing tumoral samples, we observed an increased Twist1 expression in FL compared to DLBCL, both by mRNA and protein quantification. The translocation t(14;18)(IGH;BCL2) is the cytogenetic hallmark of FL and is observed in up to 90% of this malignancy. It induces BCL2 overexpression. In DLBCL BCL2 overexpression has been demonstrated to be a prognostic factor for OS and disease-free survival (DFS) [30,31]. In this study we found a link between BCL2 and Twist1 overexpression in DLBCL (cases with BCL2 overexpression more frequently had increased Twist1 protein). Increased Twist1 in FL may correlate with BCL2 protein expression, consistent in FL, and should be associated with poor prognosis. In accordance with this hypothesis there was a tendency to shorter PFS in FL with increased Twist1 protein expression.
Although Twist1 mRNA level were low to undetectable in benign lymph nodes, immunohistochemistry demonstrated positive expression in stromal cells (micro-environment) whereas in lymphoid cells, no staining was observed. Interestingly, in FL all cases with no staining in neoplastic B cells also showed no staining stromal cells whereas in DLBCL some cases with no staining in neoplastic B cells, demonstrating Twist1 nuclear positivity in in stromal cells (data not shown). Microenvironment should play a role in Twist1 up regulation. Nuclear expression of Twist1 in stromal compartment while tumor compartment is negative has been demonstrated in breast carcinoma [37]. Authors concluded that in these cases, positive cells may correspond to 2 populations including EMT transformed neoplastic cells and stromal fibroblastic cells undergoing Twist1 activation by cytokines produced by the tumor. Twist1 expression is triggered by inflammatory-related cytokines of microenvironment and Twist1 is also expressed in follicular dendritic cells (FDC) in lymph nodes [21]. We could speculate that in lymph node and lymphoma, Twist1 may be, in part, regulated by inflammatory-related cytokines in microenvironment. Among the 8 samples of benign reactive lymph nodes, one had higher level of Twist1 mRNA and demonstrated stronger expression of Twist1 protein in stromal cells. This patient was older than other (80-years-old), had an auto-immunity history and on microscopic examination his lymph node showed heavier inflammatory lesions with increased numbers of histiocytes and hyperplastic germinal centers.
In our series, Twist1 expression in FL and DLBCL didn't correlate with adverse prognosis. No other series had studied the expression and role of Twist1 in B-cells malignancies, expected one, carried out on DLBCL, that didn't link Twist1 expression to adverse features [26]. Further investigations are needed to analyse this issue. Nevertheless, other biomarkers known to have averse prognosis in FL and DLBCL are also known to interact with Twist1.
In conclusion, our results show that Twist1 is overexpressed in FL and DLBCL. RT Q-PCR and immunohistochemistry results establish immunohistochemistry as a valuable tool in routine practice to assess Twist1 expression. In both lymphoma types, nuclear expression was observed in neoplastic B cells and stromal cells whereas only stromal cells might display positivity for Twist1 in benign conditions, suggesting a role of microenvironment in Twist1 up-regulation and B cells transformation. Although Twist1 overexpression was not directly associated with poorer prognosis in this study, Twist1 interacts with p53 and c-MYC abnormalities and NFκB and STAT3 pathways that are associated with adverse prognosis and resistance to apoptosis and to chemotherapeutic agents. As Twist1 is overexpressed in FL and DLBCL and given its interaction with other pathways involved in prognosis and drug resistance of these malignancies, further investigations are needed to characterize its function and to target it in future experimental trials.
This work was supported by an institutional grant from INSERM, and by specific grants from the CARNOT Institute CALYM and from Cancéropôle Grand Sud-Ouest to NB. NM was supported by a fellowship from the ARC foundation (fondation ARC pour la recherche sur le cancer).
Informed, written consent has been obtained.