In cell nucleus, polar and neutral lipids are responsible for the nuclear membrane and nuclear matrix fluidity. In specific sections of inner nuclear membranes that bind the active chromatin, lipid microdomains that constitute a platform for the transcription process are present.
Nuclear lipid microdomains appear as a homogenous population of closed, spherical or ovoid vesicle-like structures composed by an exact ratio of phosphatidylcholine, sphingomyelin and cholesterol. This ratio is maintained by neutral sphingomyelinase, sphingomyelin-synthase and reverse sphingomyelin-synthase activity. Interestingly, exist two pools of cholesterol in the chromatin: one called "sphingomyelin-free cholesterol fraction" and another "sphingomyelin-linked cholesterol fraction". The latest fraction is regulated by neutral sphingomyelinase and sphingomyelin-synthase activity. The organization of sphingomyelin-linked cholesterol fraction in chromatin microdomains is reported in this commentary.
Cholesterol, Nucleosome, Lipid microdomains, Liver, Phosphatidylcholine, Sphingomyelin
CHO: Cholesterol; DAG: Diacylglycerol; PC: Phosphatidylcholine; PC-PLC: Phosphatidylcholine-specific Phospholipase C; PPC: Phosphocholine; RSM-synthase: Reverse Sphingomyelin-synthase; SM: Sphingomyelin; N-SMase: Neutral-Sphingomyelinase; SM-synthase: Sphingomyelin-synthase
Cell nucleus contains both polar and neutral lipids [1] and their level is regulated by hormones [2].
Today research on the presence and role of lipids inside the nucleus is constantly evolving [3-5]. In isolated hepatocyte nuclei, polar lipids are composed by phosphatidylcholine (PC) and plasmalogen PC (PLPC), phosphatidylethanolamine (PE) and plasmalogen PE (PLPE), phosphatidylinositol (PI) and plasmalogen PI (PLPI), sphingomyelin (SM) and plasmalogen SM (PLSM), phosphatidylserine (PS) and plasmalogen PS (PLPS) [6]. Neutral lipid fraction contains cholesterol (CHO) and CHO ester [1]. 70-80% polar lipids and CHO-CHO ester are present in nuclear membrane, the remaining 20-30% resides inside the nucleus associated to the nuclear matrix, chromatin and specific intranuclear complex [1]. The most important lipids for the nucleus structure are PC, SM and CHO.
In nuclear membrane, the precise relation among these lipids is responsible for maintenance of normal fluidity in no stimulated cells. During rat liver regeneration, in which the cells are stimulated to proliferate, lipid composition and consequently fluidity of nuclear membranes modify, favoring an increase of mRNA transport that [7]. On the basis of lipids composition, the nuclear matrix seems more rigid than the nuclear membranes. The rigidity changes in relation to cell proliferation, demonstrated by fluorescent probes, reaching the maximum during DNA duplication [8]. Nuclei digested with RNAs show 84% SM decrease, thus indicating the strong relationship with RNA [1]. This was confirmed by electron microscopy using either gold SMase or antibodies against bromo-uridine (unpublished data) and by biochemical techniques isolating a nuclear fraction containing a large amount of protein, a small fraction of lipid and undigested RNA [9] and therefore called "Intranuclear Complex" (IC). Lipid analysis indicated only the presence of PC, SM and CHO. The digestion of the complex with SMase reduces SM, freeing PC and CHO, and transforms the RNAse insensitive-RNA, defined as a double strand RNA (dsRNA), into RNAse sensitive-RNA [10].
This finding demonstrates that these lipids are strongly bound to RNA and cannot be solubilized by the solvent treatment used during RNA extraction. Since N-SMase and SM-synthase activity are also present in IC, it was supposed that the digestion of SM could favor hydrolysis of RNA, whereas the synthesis of SM due to the SM-synthase could have the opposite effect, thus suggesting that the SM might represent a bridge between two RNA strands.
Moreover, incubation of RNA, isolated from hepatocyte nuclei, with low concentration of SM/CHO, results more protected RNA from RNase treatment with respect to the control [10]. It is possible to hypothesized that the lipids could facilitate the double strand formation, whereas high concentration of these lipids could play a role on the destabilization. It could explain the reason of the low increase of CHO-SM fraction with respect to dsRNA amount 24 hours after hepatectomy. Therefore, it is possible that the low increase of lipid fraction is sufficient to facilitate the formation of the high amount of dsRNA. This RNA incorporates strongly 3H-uridine, indicating that it is new-synthesized. The presence in IC of lamin B protein was the first indication that it is associated to nuclear membrane and constitutes a transcription site. During liver regeneration, the increase of new synthesized RNA is accompanied by the STAT3 activation [11]. These observations led to the hypothesis that the nuclear lipids SM-CHO and PC are organized in microdomains that could regulate nuclear function. PC-PLC activity is not present, but an equilibrium among N-SMase, SM-synthase and RSM-synthase activities is responsible for the exact ratio of PC:SM:CHO characteristic of the lipid microdomains [12].
In fact, we have demonstrated that IC actually constitutes the lipid microdomains present in the inner nuclear membrane and represents a platform for the transcription process [12]. Nuclear microdomains have a specific ratio among CHO, PC and SM that equals to approximately 1:1:1, characteristic of the lipid microdomains enriched in CHO and SM content. Therefore, the lipid composition of the microdomains is different from that of the nuclear membranes where the CHO, PC and SM ratio is 1:1.5:0.6. The electron microscopy analysis shows closed, spherical or ovoid vesicle-like structures with an average diameter in the range of 300-600 nm similar to that of microdomains isolated from cellular membrane. Lamin B is partially present in nuclear microdomains, indicating that these domains are specific parts of nuclear membrane. Differently, STAT3 is exclusively present in the microdomain fraction and represents its specific marker. PC-PLC activity is not present but an equilibrium among N-SMase, SM-synthase and RSM-synthase activities is responsible for the exact ratio of PC:SM:CHO characteristic of the lipid microdomains [12]. During liver regeneration, nuclear microdomains are characterized by an increased SM content due to higher activity of SM-synthase which increases the rigidity of the structure and permits the incorporation of 3H-uridine [12]. Therefore, the intranuclear lipid microdomains are present in the inner nuclear membrane and represent a specific section able to bind the active chromatin by acting as platform for the transcription process [13], DNA synthesis [14], vitamin D-vitamin D receptor interaction [15], and for dexamethasone action [16]. In the chromatin, SM is characterized by a high amount of saturated fatty acids, that favors the formation of Van der Waals interactions with CHO, with respect to the nuclear SM [17]. In the SMase or proteinase K digested chromatin, the amount of CHO was 1.58 times higher of that evaluated in undigested chromatin, thus suggesting the existence of two pools of CHO in the chromatin: One called "SM-free fraction", which corresponds to that detectable in untreated chromatin and another which may be measured in digested chromatin which is called "SM-linked fraction". Therefore, the CHO free level is regulated by the SM amount that, in turn, depends on SM metabolism enzyme activity [17]. Recently the presence of fat associated to the nucleosome, DNA and proteins, has been described [18]. The possible organization of PC, SM and CHO in chromatin microdomains has been considered. The chromatin was purified from nuclei of hepatocytes isolated by ultracentrifugation on sucrose gradient. Treatments with Triton X100 at 0.3% moved first the outer nuclear membrane and then the inner membrane. Subsequent ultracentrifugation allowed the formation of a precipitate containing chromatin [6]. On the precipitate, we used the technique previously used to isolate lipid microdomains from whole hepatocyte nuclei [12]. We show for the first time that the lipid microdomains rich in CHO and SM are not present only in inner nuclear membrane but also present in the chromatin. The ultramicroscopy analysis highlights the presence of a large amount of lipid microdomains in chromatin which appear as a homogenous population of closed, spherical or ovoid vesicle-like structures with an average diameter in the range of 200-300 nm. Therefore, chromatin lipid microdomains are smaller than microdomains isolated from whole nucleus but have the very similar morphology, thus indicating a specific organization inside the nucleus (Figure 1). It can be hypothesized that the lipid microdomains localized inside the nucleus might protect and drive the function of the chromatin.
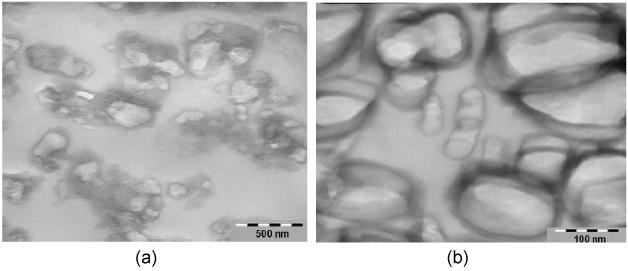 Figure 1: Sections of nucleosome lipid microdomain pellets were treated for morphology analysis by electron microscopy. Nucleosome microdomains appear as a population of closed, spherical or ovoid vesicle like structures with an average diameter in the range of 200-300 nm. a) 65000x; b) 200000x. View Figure 1
Figure 1: Sections of nucleosome lipid microdomain pellets were treated for morphology analysis by electron microscopy. Nucleosome microdomains appear as a population of closed, spherical or ovoid vesicle like structures with an average diameter in the range of 200-300 nm. a) 65000x; b) 200000x. View Figure 1