In this study, it vi cell lung carcinoma.
96 lung resection materials; it was examined for the presence of TNM (The TNM Staging System is based on the extent of the tumor (T), the extent of spread to the lymph nodes (N), and the presence of metastasis (M)) stage, smoking, age, sex, tumor differentiation and EGFR, KRAS and BRAF mutations.
Of the 96 patients included in the study, 58 were adenocarcinomas, 30 were squamous cell carcinomas (SCC), and 8 were large cell neuroendocrine carcinomas (LCNEC). While EGFR mutation was detected in 14 (24.1%) of the adenocarcinoma patients, KRAS mutation was detected in 12 (20.6%) patients. BRAF mutation was observed in 1 patient with adenocarcinoma (1.7%). KRAS mutation was observed in only 1 of the SCCs (3.3%). KRAS mutation was observed in 1 of the LCNECs (12.5%).
EGFR mutation in adenocarcinoma cases was found to be significantly associated with female gender and non-smoking status. There was no correlation between EGFR mutation and age, stage, tumor size, lymph node metastasis, distant metastasis and differentiation grade. No correlation was found between KRAS mutation and other factors. The BRAF mutation could not be assessed for association because the number of cases monitored was low.
The microarray method we used in our study provided the scanning of the cases in terms of 50 different mutations commonly identified for the EGFR gene, single chip 24 different mutations for KRAS and BRAF gene in a total of 96 cases with NSCLC (Non Squamous Cell Lung Cancer) in three different subtypes at the same time and in a short time. We think our study will be useful in the development of treatment protocols and patient selection.
Non-small cell lung carcinoma, EGFR, KRAS, BRAF, Microarray
Lung cancer is the most common type of cancer seen in males worldwide and the third most common type in females [1]. 85% of lung cancers occur in non-small cell external lung carcinomas. Small cell lung carcinomas are seen in 15%. Among non-small cell external lung carcinomas, adenocarcinoma (40%), squamous cell carcinoma (30%) and large cell carcinoma (15%) subtypes are the most common [1,2].
Over the last 10 years, the life time of patients has been increased thanks to new medicines for lung cancer treatment [3] Krizotinib, Gefitinib, Erlotinib and Monoclonal antibodies, such as nivolumab, pembrolizumab, and atezolizumab have been used to treat lung cancer for last years. In the identification of these drugs, histological type of tumor and presence of mutation is the front plan, and the drug activity depends on the biology of the tumor [4]. EGFR is the most promising oncogen in NSCLC [5]. Some specific EGFR mutations are sensitive to EGFR tyrosine kinase inhibitors [6]. Most commonly small exon 19 deletion (del 19) and exon 21-point mutation (L858R), patients were reported to respond well to EGFR tyrosine kinase treatment [7-9]. KRAS gene mutations are more common in males and adenocarcinoma-diagnosed individuals with smoking history [10]. BRAF mutation in NSCLC does not lead to worse prognosis alone, but it is sensitive to tyrosine kinase inhibitors of different classes [11-13].
The frequency of EGFR gene mutation in non-small cell lung carcinomas ranges from 58.30 to 61% for exon 10 and 36.42 to 39% for exon 21 [14-16]. The frequency of EGFR gene mutation in adenocarcinomas was from 15.5% for exon 19 and 19.3% for exon 21 [17] KRAS mutations in the histologic type of adenocarcinoma occur frequently in 25-30% of cases [1]. BRAF gene mutations are seen only 1-2% in lung cancer [18]. Tumor tissue can be heterogeneous due to its genetic structure [19]. The aim of our study is to provide the scanning of the cases in terms of 50 different mutations commonly identified for the EGFR gene, single chip 24 different mutations for KRAS and BRAF gene in a total of 96 cases with NSCLC in three different subtypes at the same time and in a short time. The target of our study is to develop treatment protocols and to be useful in patient selection.
In our study, 96 cases with non-small cell lung carcinoma diagnosed between the years 2005 and 2012 were evaluated by our department. Approval for this study was acquired from the Ethics Committee of Medical Scholl of the Akdeniz University (219209of). All cases are surgical resection material. The tissues prepared from the resection material are those fixed with 10% formalin and embedded in paraffin. The 4 micron thick sections which obtained from the patients paraffin blocks were examined by hematoxylin eosin staining. In the selection of paraffin block, samples were selected which are consisted 80% of tumor tissue. Among the cases included in the study, 58 are composed of adenocarcinoma, 30 are squamous cell carcinoma and 8 are large cell neuroendocrine carcinoma. The smoking status of the cases was determined by examining the clinical history. Ethics Committee approval has been obtained.
Histologic grade, stage, lymph node metastasis, and distant metastasis analysis according to the 2004 World Health Organization classification were determined by experienced pulmonary pathologists. Histological grade is defined as well, moderately and poorly differentiation for adenocarcinoma and squamous cell carcinoma. The staging was done according to the TNM system. For each case, primary tumor size, regional lymph node involvement, and distant metastasis were assessed.
In our study, the aim was to determine the EGFR, KRAS and BRAF gene mutations with the biofilm microarray method (Infiniti Autogenomics) in 96 cases with NSCLC. INFINITI system Autogenomics Inc. (U.S.) is an automated device based on microchip technology which can be developed by taking into consideration all the current requirements in molecular diagnosis, especially the oncology parameters applied in Molecular Pathology. The INFINITI system is a fully automated, multi-molecular diagnostic platform that uses the unique BioFilmChip Microarray for many qualitative and quantitative proteomic and genomic applications.
The Autogenomics Infiniti™ EGFR mutation screening kit was used to identify EGFR gene mutations and the Autogenomics Infiniti™ KRAS-BRAF mutation screening kit was used to identify KRAS-BRAF gene mutations. Kits are designed to identify the most common known mutations that alter protein function. The genomic regions were amplified using a multiplex polymerase chain reaction (PCR) in a thermal cycler. An enzymatic cleanup with shrimp alkaline phosphatase and exonuclease was performed after the multiplex PCR. The INFINITI Analyzer was used for allele-specific primer extension with fluorescently labeled nucleotides, capture by hybridization to the microarray, array scans and signal measurements.
Fisher, Anova, and Pearson Chi square tests were used to analyze the categorical data obtained. Kruskal Wallis test was used for comparison between the three groups. Significance value was obtained p < 0.05 in statistical analysis.
The ages of the patients included in the study ranged from 32 to 81 years. The mean age of all patients was 61.5. Of the 96 patients included in the study, 58 (60.4%) were adenocarcinomas, 30 (31.3%) squamous cell carcinomas and 8 (8.3%) large-cell neuroendocrine carcinomas. The differentiation ratios and stage of the tumors were different (Table 1 and Figure 1). 79 (82.3%) of the patients were male and 17 (17.7%) were female. Cigarette anamnesis is present in 44 of the patients. 35 of the 44 patients were smoking. Nine patients did not smoke. The smoking history of 52 patients was unknown. There was no significant relationship between smoking and histopathological types.
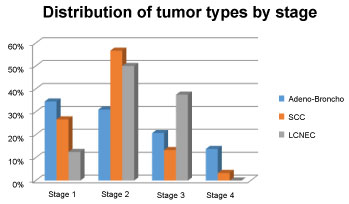 Figure 1: Distribution of tumor types by stage. View Figure 1
Figure 1: Distribution of tumor types by stage. View Figure 1
Table 1: Distribution of tumor types according to differentiation grades. View Table 1
EGFR mutations were detected in 14 (24.1%) of the patients with adenocarcinoma and 12 (20.6%) KRAS mutations were detected. BRAF mutation was observed in only 1 patient among the patients. No mutations were found in 31 patients. KRAS mutation was observed in only 1 patient (3.3%) of patients with squamous cell carcinoma, and no mutation was found in 29 patients (97.3%), KRAS mutation was observed in only 1 patient (12.5%) in patients with large cell neuroendocrine carcinoma, and no mutation was found in 7 patients (87.5%) (Table 2).
Table 2: Tumor and mutation types. View Table 2
No significant correlation was found between EGFR mutation and tumor differentiation. There was a significant difference in terms of EGFR mutation between patients with adenocarcinoma, squamous cell carcinoma and large cell carcinoma (p = 0.001). Patients with adenocarcinoma and squamous cell carcinoma were compared in terms of EGFR mutation and EGFR mutation was significantly higher than adenocarcinoma cases with squamous cell carcinoma (p < 0.001). There was no significant difference between the adenocarcinoma group and the large cell neuroendocrine carcinoma group in terms of EGFR mutation. EGFR mutation was more frequent in female patient group (p = 0.001). EGFR mutation was more frequently observed in the non-smoking group (p = 0.016). There was no correlation between EGFR mutation and the stages of patients and T, N, M values.
There was no significant relationship between KRAS mutation and age and sex. Patients with adenocarcinoma and squamous cell carcinoma were compared in terms of KRAS mutation. KRAS mutation was significantly higher in the adenocarcinoma group than in the squamous cell carcinoma group (p = 0.007). No significant difference was found between the adenocarcinoma group and the large cell neuroendocrine carcinoma group in terms of KRAS mutation. No comparison was made between the squamous cell carcinoma group and the large cell neuroendocrine carcinoma group due to inadequate sampling in terms of KRAS mutation. There was no correlation between KRAS mutation and patients' stage, T, N, M values and tumor differentiation (Table 3, Table 4 and Table 5).
Table 3: Patients characteristics, adenocarcinoma and bronchoalveolar carcinoma. View Table 3
Table 4: Patients characteristics, squamous cell carcinomas (SCC). View Table 4
Table 5: Patients characteristics, Large cell neuroendocrine carcinoma (LCNC). View Table 5
Among 96 patients, BRAF mutation was observed in a woman with only adenocarcinoma. However, this group could not be included in the statistical study due to inadequate sampling.
Adenocarcinoma, squamous cell carcinoma and large cell neuroendocrine carcinoma diagnosed in a total of 96 cases, were not seen together in two different mutations.
In males, 22% of cancer-related deaths and 15% of females are due to lung cancer [1]. The etiologic causes of lung cancer are cigarette smoking and the presence of genetic predisposition is also suggested. Current carcinogens in cigarette smoke can lead to genetic mutation by binding to specific regions of DNA. It has been shown that people with lung cancer in their family have 2-5 times more risk of cancer than other people. Damage to genes controlling cell proliferation is the main mechanism of cancer development and can occur as a result of activation of oncogenes or inactivation of tumor suppressor genes [20,21]. EGFR is a transmembrane protein with tyrosine kinase activity. KRAS protein is an activator that develops resistance to the EGFR signaling pathway. KRAS is the most frequently mutated oncogene in lung adenocarcinoma and a member of the RAS family of GTPases. It is activated through GTP binding and once GTP-bound, can trigger multiple signalling transduction pathways and impact cellular processes, about proliferation, cell survival and metabolism. KRAS is typically mutated in human cancer through missense mutations on codons that alter its protein conformation. The BRAF gene encodes a protein with serine-threonine kinase activity that is involved in the mitogen activated protein kinase intracellular signaling pathway (MAPKs). The amino acid residues that specifically encode the kinase domain of BRAF are 457-717. The activation loop of the kinase is located within the residues 59-600, which interact with the phosphate-binding loop keeping the kinase locked. Once the activation loop is phosphorylated, BRAF can also phosphorylate and thus activate the mitogen-activated 2 kinase 1 and 2 (MAP2K 1/2) signaling pathway. This gene controls cell division by effecting its activity on Kras. Gene mutation for BRAF is seen in 1-2% of lung cancers. It has been shown that the presence of KRAS and BRAF mutations in these patients lead to treatment resistance, while patients treated with the EGFR mutations respond well to treatment [11-13,22]. Although the BRAF mutation alone does not lead to poor prognosis, it is sensitive to tyrosine kinase inhibitors of different classes [12,13].
It has been reported in literature that Asian, non-smoker, female, adenocarcinoma cases with EGFR mutation respond better to treatment [23,24]. In treatment, Gefitinip, Erlotinip and monoclonal antibodies such as Cetuximab are used as tyrosine kinase inhibitors targeting the ATP binding pocket of EGFR gene and. Feng Q, et al. found more EGFR mutations in the patient group, especially those younger than 60 years, non-smokers, women and adenocarcinomas [25]. The tyrosine kinase activity of the EGFR gene is provided by exons 18-21. The most common EGFR gene mutations in the studies are the in-frame deletions in exon 19 and the L858R point mutation in exon 21 (14). The most common deletion in exon 19 is 747-leucine-749.glutamic acid deletion, which accounts for 44% of all EGFR mutations. The L858R mutation in exon 21 constitutes 41% of all EGFR mutations. The deletions and duplications in exon 20 constitute 5% [26,27]. Activating mutations in the EGFR gene affect not only the response to treatment but also the survival time at the same time. In our study, according to literature, EGFR mutation was found significantly higher in adenocarcinoma patient group than other tumor types. One of the 14 EGFR mutations detected is the L858Arg mutation at exon 21,one of them was Leu792Pro mutation in exon 19. The remaining 12 EGFR mutations were seen in exon 19 again.
EGFR and KRAS mutation association was reported to be 5% in studies conducted [28]. In one study, 99 (32.9%) of 301 patients had EGFR mutations and 14 (4.7%) KRAS mutations were detected, but no correlation was found between the two mutations [29,30]. Ohashi, et al. showed that the presence of ectopic NRAS or BRAF mutation in cells expressing the highly sensitive EGFR mutation confers resistance to EGFR tyrosine kinase inhibitor treatment [22]. The BRAF inhibitors vemurafenib, dabrafenib, and sorafenib are used for treatment in NSCLC. We did not detect KRAS and BRAF gene mutations in our study with EGFR mutations.
In the literature, the frequency of EGFR mutation in adenocarcinoma cases is reported to be 10-40% [31,32]. Yeen, et al. had studies involving 484 non-small cell lung cancer patients, EGFR mutation was significantly higher in female patients (60.6%) and adenocarcinoma (47.1%) subtypes [33]. We found EGFR mutation in 14 of the 58 adenocarcinoma cases (24.1%) in our study. EGFR mutation is associated with female gender, non-smoking status and adenocarcinoma subtype, TNM stage status is evaluated as unrelated [34-36]. We could not find any significant relationship between EGFR mutation, female gender, adenocarcinoma, non-smoker group and age, stage, tumor size, lymph node metastasis, distant metastasis and differentiation grade.
In the etiology of lung cancer it is often accused of various carcinogens including smoking. It has been observed that lung cancer seen in the non-smoking group is more likely to be younger than the smoker group, and that it is in the subtype of women and adenocarcinoma. KRAS mutation has been reported to be more common in the adenocarcinoma subtype [37]. Smoking specific nitric amines in cigarettes have been shown to induce KRAS gene mutations. Kim, et al. in a large study of adenocarcinomas, they found no association between KRAS mutation frequency and smoking [38]. Adenocarcinoma diagnosed patients with mutations in the KRAS gene has shown resistance to conventional chemotherapy, as well as high risk of recurrence and death postoperatively [11]. In adenocarcinoma histologic type, KRAS mutations are frequently seen in 5-40% and in our study, KRAS mutation was observed in 12 (20.6%) cases in 58 adenocarcinoma cases [10,39]. 12 KRAS mutations were observed in codons 12 and 13 in exon 2 which were monitored in patients with adenocarcinoma. 5 of these cases were G12 V, 4 of these cases G12C, 2 of these cases G12D and 1 of these cases were G13C subgroup observed. We could not find any significant relationship between KRAS mutation and age, sex, smokers, disease stage, tumor size, lymph node metastasis, distant metastasis and differentiation grade in our study.
In the NSCLC, high-order KRAS mutation was detected in the adenocarcinoma group based on the biofilm-based method [40]. In our study, KRAS mutation was found to be significantly higher in the adenocarcinoma group than in the SCC group. Sasaki, et al. said there was no significant relationship between BRAF mutation and gender, age, tumor stage and smoking status, but BRAF mutation was found to be significantly higher in smoker group in adenocarcinoma patients group [41]. In our study; BRAF mutation was observed in only 1 case with adenocarcinoma and there were no female patients and no smoking history.
BRAF mutations were most frequently found in the literature as point mutations in exon 15 and BRAF mutations were most frequently detected in codon 600 (V600E) by substitution of valine and glutamate amino acids [18,42]. Among our cases, BRAF V600E1 subtype mutation was observed in 1 case with adenocarcinoma.
Different molecular techniques are used in studies on mutation screening in solid tumors. The methods used to examine the genetic structure of tumor tissue should have a high mutation detection limit. With the Sanger method, DNA sequence analysis can only detect 20-25% mutation; This ratio is given as 5% for pyrosequence, and as 1% for real-time PCR. Tsiatis, et al. compared the strip test, pyrosequence and sanger sequences in a study they conducted to determine the KRAS gene mutation; as a result, the mutation detection limit of strip test was 1%, limit of pyrosequence was 5% and of limit of sanger sequence method was 20-25% [19].
Tumor tissue shows a heterogeneous structure due to its genetic. For this reason, the sensitivity of the methods used to detect even the very low mutation needs to be high and reliable [43,44]. The microarray method is more sensitive in detecting small changes in gene expression than real time PCR and Northern blot analysis [45,46]. The analyzes of the data we obtained in our study are in accordance with the results of the analyzes in previous studies and showed the reliability of our method.