Epitranscriptomic modifications of RNA, particularly N6-methyladenosine (m6A), have emerged as pivotal players in cancer biology. RNA m6A modifications dynamically regulate gene expression, influencing various aspects of RNA processing, stability, and translation. In the context of cancer, dysregulation of RNA m6A modification has been implicated in tumorigenesis, metastasis, therapy resistance, and the tumor micro environment. This review provides an in-depth exploration of the intricate relationship between RNA m6A modifications and cancer. It comprehensively outlines the enzymatic machinery responsible for m6A deposition, removal, and recognition, emphasizing how alterations in these processes contribute to oncogenesis. Global profiling techniques have revealed specific m6A patterns in various cancer types, shedding light on potential diagnostic and prognostic biomarkers. Functional insights into RNA m6A modifications underscore their impact on RNA metabolism and gene expression, influencing key oncogenic signaling pathways. Moreover, the clinical relevance of RNA m6A modifications in cancer is discussed, highlighting their potential as therapeutic targets and biomarkers for personalized medicine. This review also delves into the emerging role of RNA m6A modifications in cancer stem cells, tumor microenvironment interactions, and their cross-talk with other epigenetic marks, providing a holistic view of their contributions to cancer pathobiology. Furthermore, it discusses ongoing research, technological advancements, and future prospects in this rapidly evolving field.
In summary, this comprehensive review illuminates the multifaceted landscape of RNA m6A modifications in cancer, offering valuable insights into their diagnostic, prognostic, and therapeutic implications. Understanding the complex interplay between RNA m6A modifications and cancer biology is essential for harnessing their potential for precision oncology and novel therapeutic interventions.
N6-methyladenosine, Modifications, RNA readers and writers, RNA methylation cancer, Cancer
In recent years, the intricate world of RNA N6-methyladenosine (m6A) modifications has emerged as a captivating frontier in cancer research. These epitranscriptomic alterations, akin to the dynamic regulatory marks seen in DNA and histones, have unveiled a complex and highly regulated layer of gene expression control in cancer and various other biological processes. In this introduction, we embark on a journey into the captivating realm of RNA m6A modifications, where writers, erasers, and readers orchestrate the addition, removal, and interpretation of methyl marks on adenosine residues within RNA molecules. This epitranscriptomic code, once thought of as a mere decoration on RNA, has proven to be a pivotal regulator of gene expression, RNA stability, and translation. Today, we explore the multifaceted roles of RNA m6A modifications in cancer biology, spanning from their involvement in oncogenic signaling pathways, the regulation of cancer stem cells, and their influence on the complex tumor microenvironment. As we delve deeper into this epitranscriptomic landscape, we uncover not only the molecular intricacies but also the clinical implications that hold promise for revolutionizing cancer diagnosis and treatment, ushering in a new era of precision oncology.
RNA N6-methyladenosine (m6A) modifications represent a dynamic and reversible epitranscriptomic process that influences RNA metabolism. In recent years, the role of RNA m6A modifications has gained significant attention in the field of cancer research. These chemical alterations, catalyzed by a set of writers, erasers, and readers, have been found to play pivotal roles in regulating gene expression, RNA stability, and translation, ultimately impacting diverse aspects of cancer biology [1]. Understanding the intricate interplay between RNA m6A modifications and cancer has become essential for unraveling the molecular mechanisms underpinning oncogenesis and holds promise for the development of novel therapeutic strategies [2]. This review explores the current state of knowledge regarding the involvement of RNA m6A modifications in cancer and their potential as diagnostic markers and therapeutic targets in the fight against this complex disease [3].
RNA N6-methyladenosine (m6A) modification is a prevalent and highly conserved epitranscriptomic alteration, characterized by the addition of a methyl group to the sixth nitrogen atom of adenosine residues within RNA molecules. This chemical modification, catalyzed by a complex interplay of writer proteins (e.g., METTL3, METTL14), erasers (e.g., FTO, ALKBH5), and readers (e.g., YTHDF proteins), adds a layer of regulation to RNA, akin to DNA methylation and histone modifications in epigenetics. RNA m6A modification is dynamic, reversible, and plays a pivotal role in post-transcriptional regulation, influencing RNA processing, splicing, export, stability, and translation. It has emerged as a central player in gene expression control and is increasingly recognized for its contributions to various physiological and pathological processes, including cancer. Understanding the nuances of RNA m6A modification is fundamental to unraveling its implications in cancer biology and holds promise for novel therapeutic avenues in oncology [4].
In recent years, RNA N6-methyladenosine (m6A) modification has risen to prominence as a focal point in cancer research. This epitranscriptomic alteration, initially explored for its role in gene regulation, has gained substantial attention due to its far-reaching implications in cancer biology. Researchers are increasingly recognizing that aberrant m6A modification patterns are associated with various aspects of tumorigenesis, including the dysregulation of critical oncogenes and tumor suppressors [5]. The growing body of evidence highlights the significance of RNA m6A modification as a novel and dynamic layer of gene expression control in cancer, prompting intense investigation into its mechanistic underpinnings and potential therapeutic applications [6].
RNA N6-methyladenosine (m6A) modification, a reversible epitranscriptomic mark, is meticulously orchestrated by a group of writer proteins. Foremost among them are METTL3 and METTL14, which form a heterodimeric complex responsible for catalyzing the methyl group transfer to adenosine residues in RNA. Emerging research has revealed that these writers, along with additional cofactors, play a crucial role in fine-tuning the m6A landscape across the transcriptome. This section delves into the intricate mechanisms employed by RNA m6A writers, their structural features, substrate recognition, and regulatory functions. By understanding the precise contributions of these enzymes, we gain essential insights into the dynamic and context-dependent nature of RNA m6A modification, with significant implications for cancer and other biological processes [7].
RNA N6-methyladenosine (m6A) modification, a pivotal epitranscriptomic process, relies on a specialized group of enzymes for its precise installation. Central to this machinery are the writer proteins, primarily METTL3 and METTL14, which form a stable heterodimeric complex. Together with additional cofactors, they catalyze the methyl group transfer from S-adenosyl methionine (SAM) to adenosine residues in RNA. The METTL3-METTL14 complex functions as the core methyltransferase, recognizing specific RNA sequence motifs and ensuring site-specific m6A modification [7]. This section provides an in-depth exploration of the structural features, mechanistic insights, and regulatory aspects of these enzymes, highlighting their pivotal role in shaping the epitranscriptomic landscape. Understanding the enzymatic machinery behind m6A methylation is essential for unraveling its implications in gene expression regulation, RNA processing, and its significance in diverse biological contexts, including cancer [1].
The precise orchestration of RNA N6-methyladenosine (m6A) modification is essential for normal cellular functions, but in cancer, this tightly regulated process often goes awry. Dysregulation of m6A modification patterns is a hallmark of various malignancies and is associated with oncogenic transformation. Aberrant expression or activity of m6A writers, erasers, and readers can disrupt the delicate balance of RNA methylation, leading to widespread alterations in gene expression, RNA stability, and translation. These disruptions contribute to key aspects of cancer biology, including the unchecked growth of tumor cells, metastasis, therapy resistance, and immune evasion [8]. Understanding the molecular intricacies of m6A dysregulation in cancer is pivotal for developing targeted therapeutic strategies and diagnostic approaches in the fight against this complex disease [9].
RNA m6A erasers, prominently represented by FTO (Fat Mass and Obesity-associated protein) and ALKBH5 (AlkB Homolog 5), constitute a critical facet of the intricate epitranscriptomic machinery responsible for governing RNA N6-methyladenosine (m6A) modifications. These enzymes possess the unique capability of erasing m6A marks, effectively demethylating adenosine residues within RNA molecules. By doing so, RNA m6A erasers introduce dynamicity into the RNA modification landscape, allowing for precise post-transcriptional control [7]. Their regulatory functions encompass a broad spectrum of RNA processes, including degradation, translation, and stability, thereby exerting significant influence over gene expression patterns [10]. Dysregulation of m6A erasers has emerged as a key feature in various diseases, notably cancer, where it can lead to pervasive alterations in the transcriptome, contributing to oncogenic transformation, metastasis, and therapeutic resistance. The study of RNA m6A erasers continues to illuminate the intricate interplay within the epitranscriptomic code and holds potential for innovative therapeutic strategies targeting epitranscriptomic marks in disease contexts [11].
RNA m6A readers, a diverse group of proteins that recognize and interpret N6-methyladenosine (m6A) modifications in RNA, play a pivotal role in the epitranscriptomic code. These readers, exemplified by YTH domain-containing proteins (YTHDFs and YTHDCs), selectively bind to m6A-modified transcripts, thereby acting as molecular switches that dictate the fate of methylated RNAs [12]. By recruiting various downstream effectors, m6A readers impact multiple aspects of RNA metabolism, including mRNA splicing, export, stability, and translation. Their intricate regulatory functions contribute to the fine-tuning of gene expression, and their misregulation can have profound consequences in diseases, including cancer. Understanding the diverse and context-dependent roles of RNA m6A readers is instrumental in deciphering the epitranscriptomic code and holds promise for uncovering novel therapeutic avenues in cancer and other m6A-associated pathologies [13,14] (Figure 1).
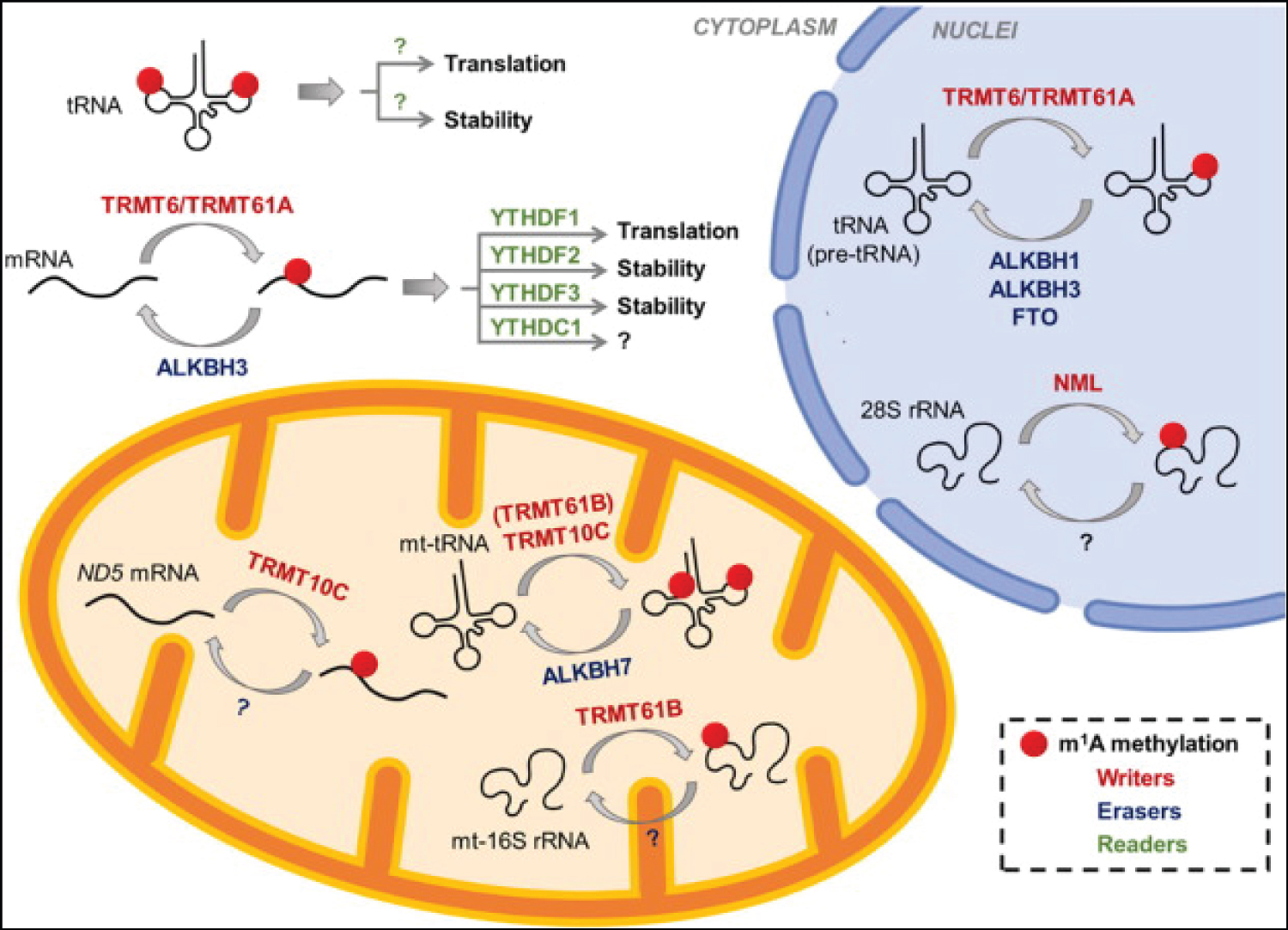 Figure 1: RNA m6A readers, writers and markers (Li, J., Zhang, H., & Wang, H. (2022). N1-methyladenosine modification in cancer biology: Current status and future perspectives. Computational and Structural Biotechnology Journal).
View Figure 1
Figure 1: RNA m6A readers, writers and markers (Li, J., Zhang, H., & Wang, H. (2022). N1-methyladenosine modification in cancer biology: Current status and future perspectives. Computational and Structural Biotechnology Journal).
View Figure 1
Global profiling of RNA N6-methyladenosine (m6A) modifications has emerged as a transformative approach in cancer research, shedding light on the extensive landscape of epitranscriptomic alterations in various malignancies. Utilizing cutting-edge high-throughput techniques such as MeRIP-seq (Methylated RNA Immunoprecipitation Sequencing) and m6A-seq, researchers have uncovered specific m6A modification patterns across the transcriptome of cancer cells. This comprehensive mapping not only identifies individual m6A-modified transcripts but also provides insights into their regulatory roles and functional consequences. Such global profiling efforts have unveiled novel m6A-regulated genes, pathways, and networks, advancing our understanding of the molecular mechanisms underlying tumorigenesis. Furthermore, this research has immense potential for biomarker discovery, offering valuable diagnostic and prognostic insights, as well as therapeutic targets tailored to the unique m6A modification profiles associated with different cancer types [15-17].
Functional implications of RNA N6-methyladenosine (m6A) modifications are at the forefront of epitranscriptomic research, as they govern a myriad of post-transcriptional processes with profound consequences for gene expression regulation. M6A modifications act as molecular tags on RNA molecules, orchestrating RNA processing, splicing, export, translation, and stability. These modifications fine-tune gene expression, allowing cells to rapidly adapt to changing environmental conditions and developmental cues [18]. Dysregulation of m6A modification patterns in diseases, particularly cancer, can lead to significant alterations in gene expression profiles, affecting critical oncogenes and tumor suppressors. As we unravel the functional intricacies of RNA m6A modifications, we gain a deeper understanding of their roles in cellular homeostasis and disease pathogenesis, opening new avenues for targeted therapeutic interventions and precision medicine approaches in cancer and beyond [19].
RNA N6-methyladenosine (m6A) modifications intersect with pivotal oncogenic signaling pathways, forging intricate connections that influence cancer progression and treatment responses. This crosstalk involves both direct and indirect interactions, with m6A modifications affecting the activity and expression of key pathway components. For instance, m6A modifications can impact the expression of growth factors, kinases, and transcription factors, thus modulating the PI3K-AKT, MAPK/ERK, Wnt/β-catenin, and other pathways critical for tumorigenesis. Additionally, m6A modifications can fine-tune the expression of genes involved in DNA repair, apoptosis, and cell cycle regulation, thereby influencing the sensitivity of cancer cells to therapies targeting these pathways [20]. Understanding the complex interplay between RNA m6A and oncogenic signaling pathways is pivotal for unraveling the molecular mechanisms underpinning cancer pathogenesis and therapy resistance, ultimately offering novel therapeutic strategies and predictive biomarkers in the realm of precision oncology [21].
Interactions between RNA N6-methyladenosine (m6A) modifications and key cancer pathways lie at the heart of cancer biology, revealing a web of regulatory connections that orchestrate tumorigenesis. These epitranscriptomic marks play a multifaceted role in influencing oncogenic signaling pathways, such as the PI3K-AKT, MAPK/ERK, and Wnt/β-catenin cascades. M6A modifications can directly impact the expression and activity of critical pathway components, including ligands, receptors, kinases, and transcription factors, leading to alterations in cell proliferation, survival, and metastatic potential. Moreover, m6A-mediated regulation extends to genes involved in DNA repair and apoptosis, significantly shaping cancer cells' response to therapy. These intricate interactions underscore the complexity of RNA m6A's involvement in cancer biology and highlight its potential as a nexus for targeted therapeutic interventions aimed at disrupting oncogenic signaling networks [22,23] (Figure 2).
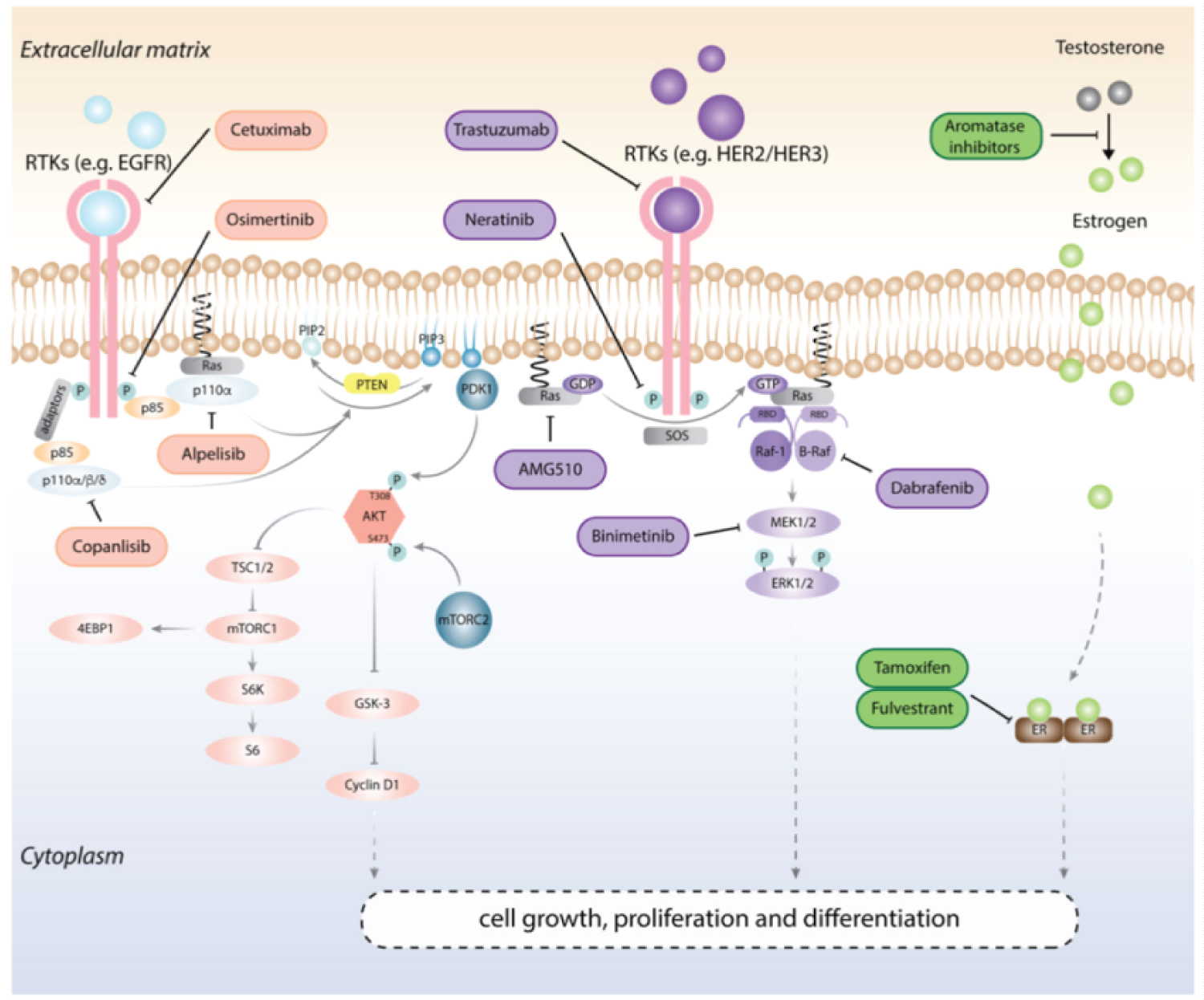 Figure 2: Cancer pathways in context with signaling and RNA m6A (Chen, M., & Wong, C. M. (2020). The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Molecular cancer, 19, 1-12).
View Figure 2
Figure 2: Cancer pathways in context with signaling and RNA m6A (Chen, M., & Wong, C. M. (2020). The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Molecular cancer, 19, 1-12).
View Figure 2
The clinical relevance of RNA N6-methyladenosine (m6A) modifications in cancer is becoming increasingly evident, holding promise for diagnostic, prognostic, and therapeutic applications. Emerging research has linked dysregulated m6A modification patterns to various cancer types, highlighting their potential as diagnostic biomarkers. The detection of specific m6A modification profiles in tumor tissues or circulating RNA can aid in cancer diagnosis and stratification. Additionally, these modifications offer valuable prognostic information, with certain m6A patterns serving as indicators of disease aggressiveness, metastatic potential, and treatment outcomes [24]. Furthermore, the targeting of RNA m6A modifications, through the development of small molecules or RNA-modifying enzymes, represents an innovative avenue for precision cancer therapy [25]. By modulating m6A modification levels in specific transcripts, it becomes possible to selectively inhibit oncogenic pathways, enhance immune responses, or sensitize cancer cells to existing therapies. Overall, the clinical relevance of RNA m6A modifications in cancer underscores their potential to transform the landscape of cancer diagnosis and treatment, ultimately improving patient outcomes and personalized care [26].
The diagnostic and prognostic potential of RNA N6-methyladenosine (m6A) modifications in cancer is increasingly recognized as a promising avenue for improving patient care. M6A modification profiles, unique to different cancer types and even subtypes, can serve as molecular signatures that aid in accurate cancer diagnosis and classification. These profiles can be detected in tumor tissues, blood, or other biofluids, offering non-invasive and precise diagnostic tools. Moreover, the dynamic nature of m6A modifications provides valuable prognostic insights. Specific m6A patterns have been associated with clinical outcomes, including survival rates and disease progression, enabling better risk stratification and treatment decisions. As our understanding of the relationship between m6A modifications and cancer advances, harnessing their diagnostic and prognostic potential holds great promise for tailoring individualized treatment strategies and ultimately improving patient care and outcomes [26].
Biomarker development and their integration into clinical trials represent a crucial step in translating RNA N6-methyladenosine (m6A) modifications' potential into clinical practice for cancer management. M6A modification patterns, when validated as reliable biomarkers, can help identify patient subgroups likely to respond to specific treatments. Clinical trials that incorporate these biomarkers as part of their design enable a more targeted and personalized approach to cancer therapy. By selecting patients based on their m6A modification profiles, these trials aim to improve treatment outcomes, reduce adverse effects, and optimize therapeutic strategies. The development of m6A-based biomarkers, coupled with their integration into clinical trials, holds the promise of ushering in a new era of precision oncology, where treatments are tailored to individual patients' molecular characteristics, ultimately enhancing therapeutic efficacy and patient care [27].
Therapeutic targeting of RNA N6-methyladenosine (m6A) in cancer is an emerging and promising strategy that holds significant potential for improving cancer treatment outcomes. Given the pivotal role of m6A modifications in regulating oncogenic pathways and gene expression, researchers are actively exploring various approaches to modulate these epitranscriptomic marks for therapeutic purposes [28-31].
1.Small Molecule Inhibitors
2. RNA-Targeting Therapies
3. Immunotherapeutic Approaches
4. Epitranscriptomic Editing
5. Combination Therapies
RNA N6-methyladenosine (m6A) modification plays a critical role in the regulation of cancer stem cells (CSCs), a subpopulation of cancer cells known for their self-renewal and tumorigenic properties. M6A modifications are intricately involved in maintaining the balance between self-renewal and differentiation in normal stem cells, and this regulatory mechanism is often hijacked in CSCs to promote their survival and proliferation [32]. Here are some key points regarding RNA m6A modification in cancer stem cells:
• Maintenance of CSC identity: M6A modifications are involved in sustaining the unique identity of CSCs. They regulate the expression of genes associated with stemness, which is crucial for CSC self-renewal and pluripotency [32].
• Enhanced tumorigenicity: Dysregulated m6A modification patterns in CSCs can lead to increased tumorigenicity. Aberrant m6A regulation may promote CSC survival and resistance to therapies, contributing to cancer recurrence and metastasis [29].
• Regulation of signaling pathways: M6A modifications impact signaling pathways that are critical for CSC function, such as the Wnt/β-catenin and Notch pathways. Dysregulation of m6A marks on key transcripts can enhance the activation of these pathways in CSCs [33].
• Therapeutic implications: Targeting m6A modifications in CSCs is a promising strategy for cancer therapy. Modulating the epitranscriptomic landscape of CSCs can sensitize them to conventional treatments and reduce their tumorigenic potential [34].
• Diagnostic and prognostic value: Characterizing the m6A modification profiles in CSCs may have diagnostic and prognostic value. Specific m6A patterns in CSC-related transcripts could serve as biomarkers for identifying aggressive tumor subtypes or predicting treatment responses [34,35].
The relationship between RNA N6-methyladenosine (m6A) modifications and the tumor microenvironment (TME) is an emerging area of research with significant implications for cancer biology and therapy [36]. Here are some key points regarding the interplay between RNA m6A and the TME:
• Immune response regulation: M6A modifications can influence the expression of immune-related genes within the TME. They may affect the recruitment, activation, and function of immune cells, including T cells, B cells, and myeloid cells, which play a crucial role in the anti-tumor immune response [37].
• Tumor-Infiltrating lymphocytes (TILs): M6A modifications can impact the RNA stability and translation of cytokines, chemokines, and immune checkpoint molecules in cancer cells. This can shape the TME and the composition of tumor-infiltrating lymphocytes (TILs), affecting immune surveillance and evasion [38].
• Inflammatory response: Dysregulated m6A modifications in cancer cells can lead to chronic inflammation within the TME, contributing to tumor progression and therapy resistance. Inflammatory signals can, in turn, influence m6A writer and eraser expression, creating a feedback loop [39].
• Therapeutic implications: Understanding the impact of m6A modifications on the TME offers therapeutic opportunities. Modulating the m6A epitranscriptomic landscape may enhance the effectiveness of immunotherapies, as well as other treatments aimed at reshaping the TME [40].
• Biomarker potential: Profiling m6A modifications within the TME can provide valuable biomarkers for predicting immune responsiveness, treatment outcomes, and patient prognosis. These biomarkers may guide personalized treatment strategies. Immune Responses and Tumor-Infiltrating Cells [41].
Future directions and emerging research in the field of RNA N6-methyladenosine (m6A) modifications are poised to unlock new insights into the complexities of epitranscriptomics and its implications for various biological processes, particularly in the context of cancer. Here are some key areas of focus and research trends:
• Dynamic RNA modifications: Investigating the dynamics of m6A modifications, including their reversible nature and temporal regulation, will continue to be a priority. Researchers aim to develop real-time tracking methods to monitor changes in m6A status in response to cellular stimuli and during disease progression [42].
• M6A readers, writers, and erasers: Deeper exploration of the roles and regulation of m6A readers, writers, and erasers will uncover novel functions and regulatory mechanisms. Understanding their context-dependent activities and interactions with other RNA-binding proteins is crucial [42].
• Functional genomics: Comprehensive functional genomics studies will provide insights into the specific roles of m6A modifications in gene expression regulation, RNA processing, and disease pathogenesis. Genome-wide screens and high-throughput assays will continue to uncover m6A-regulated pathways and targets [43].
• Single-Cell analysis: Single-cell RNA sequencing and epitranscriptomic profiling will allow researchers to dissect the heterogeneity of m6A modifications within complex tissues and tumor microenvironments, shedding light on cell-specific roles in health and disease.
• Therapeutic strategies: The development of therapeutics targeting m6A modifications, including small molecules and RNA-based therapies, will advance. Combining these approaches with existing cancer treatments holds promise for improving therapeutic outcomes [44].
• Clinical translation: Validating the clinical relevance of m6A modifications as diagnostic, prognostic, and predictive biomarkers will be crucial. Clinical trials assessing the efficacy of m6A-targeted therapies in cancer and other diseases will continue to evolve [45].
• Epitranscriptomics in disease: Expanding our understanding of how dysregulated m6A modifications contribute to various diseases beyond cancer, such as neurological disorders and metabolic diseases, will be a growing area of research.
• Technology advancements: The development of innovative technologies for studying RNA modifications, such as nanopore sequencing and mass spectrometry, will enhance our ability to accurately map and quantify m6A sites across the transcriptome [45].
In conclusion, RNA N6-methyladenosine (m6A) modifications have emerged as a dynamic and multifaceted epitranscriptomic regulatory mechanism with far-reaching implications in cancer and beyond. Our understanding of m6A modifications has expanded significantly, from their roles in gene expression regulation and RNA metabolism to their involvement in oncogenic signaling, cancer stem cells, the tumor microenvironment, and therapeutic targeting. As research in this field continues to advance, several clinical implications become evident: Profiling m6A modification patterns has the potential to yield valuable diagnostic and prognostic biomarkers for cancer and other diseases. Specific m6A signatures may guide personalized treatment strategies and enhance patient care.
Targeting m6A modifications in cancer represents a promising avenue for therapy development. Small molecules, RNA-based therapies, and epitranscriptomic editing tools hold potential for disrupting oncogenic pathways and enhancing treatment efficacy. Incorporating m6A-related information into precision medicine approaches can lead to more tailored and effective treatments. By considering m6A modification profiles, clinicians may better predict therapeutic responses and select optimal treatment strategies. Modulating m6A modifications in the tumor microenvironment can potentially enhance the efficacy of immunotherapies. This approach may sensitize cancer cells to immune attack and overcome resistance mechanisms.
Expanding research into m6A modifications in other diseases, such as neurological disorders and metabolic conditions, may reveal novel biomarkers and therapeutic targets. Ongoing research into the dynamics, regulation, and functional consequences of m6A modifications will continue to uncover new insights, shaping our understanding of gene regulation and disease mechanisms. In essence, RNA m6A modifications represent a burgeoning frontier in molecular biology with profound implications for translational medicine. They hold the promise of transforming our approach to disease diagnosis, prognosis, and therapy, offering new avenues for precision medicine and improving patient outcomes. As our knowledge in this field deepens, we anticipate innovative strategies and clinical applications that will revolutionize the landscape of healthcare.
Afrasiyab: Author of review article RNA m6A modifications and Cancer; Sadaf Ayaz: Headings covering the “readers, erasers and writers of RNA”; PeilinZheng: Global profiling, headings covering the therapeutics and biomarkers; Shoujun Huang: Correspondence author covered headings of future goals and conclusion.
All listed authors have reviewed the final version of manuscript, approved its submission and have no conflicts of interests.
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 32070616 and 82170794), Innovation and Technology Bureau of Longhua, Shenzhen (Key Laboratory of Immunodiagnostics, and Shenzhen Municipal Science and Technology Innovation Committee Project (JCYJ20190807150619047).