COVID-19 is a highly infectious disease caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Although the exact diagnosis, clinical and treatment features for SARS-CoV-2 infection have not been determined yet, the accumulation of knowledge is increasing day by day. However, there is still insufficient information about the cellular and molecular regulation of SARS-CoV-2. The development of miRNA targeting anti-viral therapies in the literature has attracted great interest and have been considered as promising biomarker(s) and novel target(s) for therapeutic approaches. MicroRNAs are powerful post-transcriptional regulators of the gene expression. They have been found as important modulators of viral infections that can play an important role in the treatment. MiRNA expression profiles are used as molecular markers for the classification, diagnosis, and progression of viral biomarkers. According to the mİRBase, database of published miRNA sequences, there are 38589 miRNA entries found in many species recently. It is believed that there are many more miRNAs to be discovered for controlling of gene expression. The conclusion to be drawn from the studies is that potential miRNA subsets of patients can be targeted to treat COVID-19. This review summarizes potential miRNAs that can be targeted for the further therapies of SARS-CoV-2 infections. In this article, a general review of current studies concerning the function of miRNAs in SARS-CoV-2 infection, and available therapeutic prospects to mitigate the burden of viral infections were presented. Also, future directions of miRNA studies for SARS-CoV-2 infection are also pointed aiming therapeutic approaches and personalized therapies for the infected patients.
SARS-CoV-2, COVID-19, Viral miRNAs, Host miRNAs, miRNA therapeutics
Coronaviruses are a group of single stranded RNA viruses (ssRNA) with viral spike proteins on their surface [1]. SARS-CoV-2 is a strain of severe acute respiratory syndrome-related coronavirus (SARSr-CoV), belongs to the broad family of coronaviruses [2]. Other coronaviruses are capable of causing illnesses including common cold and more severe diseases such as Middle East respiratory syndrome (MERS). After 229E, NL63, OC43, HKU1, MERS-CoV, and SARS-CoV, SARS-CoV-2 is the seventh known coronavirus to infect people [3]. Considering today's mortality rates, SARS-CoV-2 requires novel and in-depth experimental studies to encourage new strategies for the management of this pandemic.
The coronaviruses' genomic structure contains number of open reading frames (ORF) and four structural proteins; surface glycoprotein (S), membrane (M), envelope (E), glycoprotein and nucleocapsid phosphoprotein (N) as shown in Figure 1. The surface structure has the S proteins that creates spikes to bind to host receptors. These S proteins have been imaged at the atomic level with cryogenic electron microscopy [4]. S protein comprise of S1 and S2 subunit. S1 subunit catalyzes the attachment, determines virus-host range and cellular orientation and S2 subunit catalyzes the membrane fusion of the host cell and the virus [5].
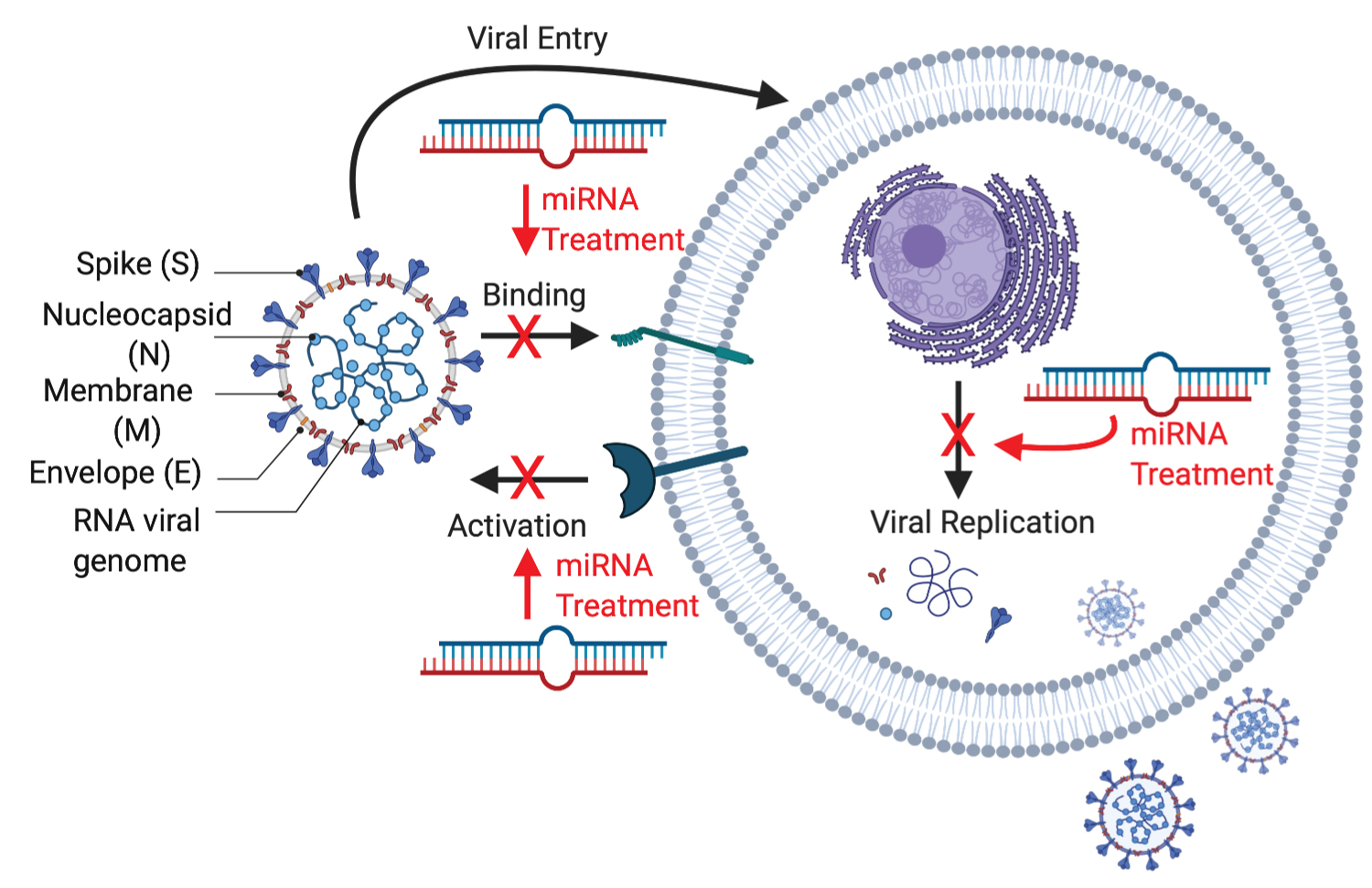 Figure 1: SARS-CoV2 structure and potentialtarget sites of miRNA therapeutics. Created with BioRender.com.
View Figure 1
Figure 1: SARS-CoV2 structure and potentialtarget sites of miRNA therapeutics. Created with BioRender.com.
View Figure 1
The M protein has structure of three transmembrane domains and to bind the nucleocapsid. The E protein is important for viral pathogenesis and virus recognition. The N protein binds the RNA of the virus via two different domains and holds the viral genomic structure [6].
SARS-CoV-2 RNA structure has incorporation of a polybasic cleavage site, therefore, it has characteristics of higher potential of pathogenicity and transmissibility than other viruses. It also has been predicted that SARS-CoV-2 may use extracellular matrix metalloproteinase inducer to facilitate cell entry which makes them more infectious to humans [7].
SARS-CoV-2 has a genomic size of 29,903 nucleotides with a 5 ʹ-cap structure and 3 ʹ-poly A tail single stranded RNA (NCBI genome ID:86693) [8]. Based on the studies, it has been shown that SARS-CoV-2 has a higher affinity to human ACE2 than the original SARS virus strain [9]. Transmembrane protease, serine 2 (TMPRSS2) is an enzyme and essential for cellular entry of SARS-CoV-2 [10]. The S protein of the virus is opened by TMPRSS2, a fusion peptide is exposed in the S2 subunit [5].
Around the virion, endosome structure is formed after the fusion to isolate the virus from the host cell. The viral RNA is released into the cell to induce the cell to produce copies of virus to infect more host cells. According to Hoffman, TMPRSS2 inhibitor can be used to block the entry and might constitute a treatment option, which is being investigated as a possible treatment for COVID-19 [10]. MiRNAs have been revealed to play crucial roles in a complex network of interaction between the virus and infected cells and they can have great therapeutic potential as shown in Figure 1 [11].
Micro RNAs or mRNA-inhibiting RNAs (miRNAs) are non-protein coding RNA molecules (17–24 nt) that bind to the target mRNA and regulate the gene expression by suppressing the protein conversion process or disrupting the mRNA, providing high selectivity with a single gene or several target genes interactions. MiRNAs have crucial roles in homeostatic processes such as cellular proliferation, differentiation or death. In recent studies including the viral infections, miRNAs have emerged as important regulators of viral infections to suppress the gene expression by targeting cellular or viral RNAs during infection. They are widely found in plants, animals and some viruses including SARS-CoV-2 [12].
Cellular miRNAs can inhibit the translation of the viral genome to prevent virus replication. Also, these miRNAs can also stabilize the viral RNA within specific cells and/or tissues [13]. According to the WHO initiative Battle against Respiratory Viruses, novel anti-viral therapeutic approaches are needed to treat respiratory viral infections [14].
In general, 3′ untranslated region (3′ UTR) of target mRNAs are the target sites of miRNAs to induce mRNA degradation and translational repression. On the other hand, gene activating role of some miRNAs have been showed with increasing number of evidences. They can interact with the 5′ UTR and gene promoters to activate the gene expression [15].
Infected host cell miRNAs can facilitate the viral immune evasion by targeting some important immune responses. Host miRNAs can also be induced as a pro-viral factor by inhibiting host immune pathways. These miRNAs can downregulate different Toll-Like Receptors (TLRs) signaling which are the primary stimulatory molecules for producing interferons and other inflammatory cytokines as an antiviral response [16].
Up to date, viral gene regulations have been targeted by natural miRNAs or created synthetic small interfering RNAs (siRNAs). In some cases, positive results were obtained in in vitro studies but it was not applicable in in vivo studies because of the lack of evidence that miRNA and siRNA do not have side effects in humans or experimental animals. Considering the information obtained to date in the literature of SARS-CoV-2, there are a very limited number of studies on the role of miRNAs in disease control and most of the studies have not been established experimentally [17].
Although most of the SARS-CoV-2 and miRNA related studies have been made in silico analysis, there are some studies that present potential miRNA gene regulations experimentally. Researchers have demonstrated that microRNAs, which are most competent against SARS-CoV-2, are effective in more than 10 target regions and are ultimately the most competent in fighting the virus. They suggested that SARS-CoV-2 infected patients may have the same microRNAs indicating that effective treatments or vaccines can have a broad effect. The most associated miRNAs with COVID-19 disease were found as miR-15b-5p, miR-15a-5p, miR-548c-5p, miR-548d-3p, miR-409-3p, miR-30b-5p and miR-505 [18].
In another experimental study, thrombotic and thromboembolic complications have been demonstrated to play a critical role in the clinical outcome of COVID-19. Exosomal miRNAs are functionally involved in a number of physiological and pathological processes. Due to their small size, natural products of the body cells exosomes are an excellent delivery system for anti-disease miRNAs in therapeutic tools. Exosome encapsulated miRNAs are highly stable in blood circulation since the exosomes can protect miRNA structure against RNase-mediated degradation. In this study, circulating exosomes were isolated from patient serum samples and exosomal miRNA levels were measured. D-dimer is a fragment (two D fragments of the fibrin protein) of fibrin degradation present in the blood after a blood clot is degraded and it can be determined by a blood test to help diagnose thrombosis. Dramatically, exosomal miR-424 was found to be significantly upregulated compared to patients in the high d-dimer group. However, exosomal miR-103a, miR-145 and miR-885 were found to be downregulated in the high d-dimer group [19].
It has been shown that the patients with COVID-19 that don't respond to the tocilizumab treatment, the levels of miR-146a-5p were decreased in their serum. MiRNAs in the blood, such as miR-146a-5p, -21-5p, and -126-3p can be potential biomarkers to provide a molecular link between inflammation and the COVID-19 [20]. In an experimental study on rat and human derived cardiomyocytes, it has been shown that both ACE2 mRNA and protein levels are inhibited by miR-200c, which can also be the potential target of the miRNA therapeutics in cardiomyocyte-affected infections [21]. In another cell culture experiment, Caco-2 cells were transfected with miRNA(s) and silencing of Tmprss2 with maximum gene suppression by hsa-miR-32 were proved [22].
In a study evaluated the microRNAs that specifically target TMPRSS2, through a bioinformatic approach, miR-98-5p was identified as a suitable miRNA candidate. It has been experimentally validated that miR-98-5p can be a regulator of TMPRSS2 transcription in two different human endothelial cell types, derived from the lung and the umbilical vein [23].
MiR‐6501‐5p and miR‐618 was found higher in the SARS-Co-2 infected patients' peripheral blood than that of the healthy donors. Moreover, miR‐627‐5p was found as the most downregulated miRNA in peripheral blood samples of the patients compared to that of the controls. The expression of miR‐183‐5p, miR‐627‐5p, and miR‐144‐3p were found to be reduced by more than 1.3‐fold in infected patients compared to healthy donors' blood samples [24]. In a different study, serum concentration of miR-21, miR-155, miR-208a and miR-499 were found to be dramatically increased in SARS-CoV-2 infected patients compared to healthy controls [25].
Computational analysis of the miRNAs on virus infection is helpful in developing new drugs to attack SARS-CoV-2 infection. To elucidate this, researchers use the computational and in silico analysis to find the interaction network of SARS-CoV and human miRNAs [26]. The results from the studies point out that coronavirus infections can be induced or inhibited by certain miRNA expressions in host cells. Therefore, host cell miRNAs could be the best option for exploration of miRNA-based therapies for coronavirus diseases.
Mainly, miRNA and COVID-19 related studies have focused on computational and in silico analysis in the literature. In a computational study, miR-4778-3p, miR-6864-5p and miR-5197-3p were identified as the most effectively interacting miRNAs with the RNA of the coronaviruses. Use of miRNAs specific to the SARS-CoV-2 RNA genome to protect lung cells from inflammation is suggested. MiRNA-target prediction via bioinformatics analysis revealed that miR-4778-3p, miR-6864-5p and miR-5197-3p were identified as miRNA candidates with the most effectively interacting with the RNA of the coronaviruses [27]. MiRNA-based therapy could be proposed for SARS-CoV-2 treatment through the viral genome suppression. In this line, a comparative computational viral genome analysis showed that six host miRNAs, including miR-101, miR-126, miR-23b, miR-378, and miR-98 can be considered as anti-viral miRNAs which could suppress SARS-CoV-2 nonstructural protein (nsp), N and S glycoprotein as a target [28].
Complementary miRNA (cc-miRNA) that could specifically inhibit the translation of viral proteins by strong interactions with the viral genome were also proposed. They identified the cc-miR in the RNA of the coronaviruses by using miR- 6864-5p, miR-4778-3p, and miR-5197-3p. These miRNAs associated with the RNA structure of the virus to inhibit the genomic replication. Synthesis viral proteins and reproduction of viral genome were suggested to be inhibited by these cc-miRNAs. It has been also suggested that cc-miRs could be used by being fused with exosomes or other biological vesicles to be transferred within the lung through inhalation. By this way, cc-miRNAs within the blood can inhibit the viral infection in the bloodstream and the organs [29].
Potential miRNAs were predicted according to RNA base-pairing and miR-1307-3p and miR-3613-5p were predicted as biomarkers to prevent virus replication via targeting 30-UTR of the host cell genome [30]. It also has been reported that immune response and cytoskeleton organization processes are regulated by the miRNAs. From the analysis, hsa-miR-4661-3p was predicted to target SARS-CoV-2 (S gene) [31].
In a comprehensive computational study, the different target sites of SARS-CoV-2 including membrane (M), envelope (E), nucleocapsid (N), replication and spike protein (S) and potential host (human) miRNAs were identified. They have shown that except envelope (E) protein (targeted by miR-3672) and ORF6 (targeted by miR-190a-5p) all viral genes can be targeted by more than one host miRNAs. They considered that the functions of SARS-CoV-2 coding genes might share functional similarity with SARS-CoV, therefore, potential miRNAs were listed. The target sites of the potential miRNAs on viral genes are listed in Table 1. They also suggested that increase in the level of host miRNAs targeting viral genes would hinder viral entry and replication [32].
Table 1: MiRNA studies related to SARS-CoV-2 and their functions, outcomes/potential targets. View Table 1
The genomic mutations on targets of virus, host miRNA and viral miRNA might contribute to the attenuated phenotype of SARS-CoV-2 during its evolution. The virus the altered genome might trigger the high level of immunity response and the cytokine storm. Moreover, the mutations may result in enhance in invasion of the virus through attracting different batches of host miRNAs [30]. However, the in-silico studies of different potential miRNAs of SARS-CoV-2 infection require experimental validation in vitro and in vivo models in further investigations.
These miRNAs can also induce other mechanisms to regulate the changes in host cell activity or the virus replication [33]. Virus protects itself to struggle and avoid host immune system. Viral miRNAs and their biological functions have become a research hotspot in recent years. Therefore, miRNAs of the SARS-CoV-2 need to be interpreted to understand its action mechanism for basic research and drug development [34].
Many viral miRNAs were found to assist in their overall pathogenesis by regulating host gene expressions [35]. In a computational study, the similarities between the SARS-CoV-2 genome and regulatory human miRNAs were predicted and 7 miRNAs (miR 8066, 5197, 3611, 3934-3p, 1307-3p, 3691-3p, 1468-5p) were determined [26]. SARS-CoV-2 encoded miRNAs can target several host genes to regulate (suppress or escape) the host's immune surveillance. Host cell's Wnt, TNF-alpha, ErbB, mTOR, VEGF, TGF-beta and MAPK signaling pathways, T cell-mediated immunity, autophagy, FGF receptor binding are particularly targeted by SARS-CoV-2 that affect heart and brain development, and insulin signaling. In a computational study, Ca2+ signaling pathway, which is considered as crucial activator of many signaling pathways is found to be targeted by the SARS-CoV-2 miRNAs [16]. Virus-encoded miRNA, MR147-3p, is predicted to enhance the expression of TMPRSS2 which result in strengthening SARS-CoV-2 infection in the gut [30]. In another study, three miRNA precursor sequences of the SARS-CoV-2 were identified by in silico analysis. The viral genome was compared with other genes by using a variety of prediction tool [35].
Although in silico analysis of virus or host miRNAs would provide cost and time benefits to figure out the underlying molecular mechanisms of SARS-CoV-2 infection, experimental validation is needed in further investigations.
The miRNAs can be incorporated into exosomes or other vesicles and delivered to the blood or lung by inhalation. After the delivery, the miRNAs can suppress the reproduction of the virus in the blood and in target organs. Up to date, there is no available therapeutics to prevent or inhibit miRNAs in viral infections. Synthetic miRNAs that are carried by liposomes have been generated in recent studies. Artificial miRNAs were generated and mononuclear cells of peripheral blood were transfected with liposomal carriers [36]. MiR-34 (MRX34) is the first developed synthetic RNA for the treatment of advanced hepatocellular carcinoma [37]. Natural RNA molecules are used in many studies since they have no side effects in in-vivo and can diffuse in the cells without the transmission vehicles or transfecting tools. Since RNA molecules are not much alive, they can be transported via lipid nanoparticles as performed in phase-I clinical trial in SARS-CoV-2 infected patients. mRNA-1273 is transported via lipid nanoparticles in SARS-CoV-2 infected patients [38]. Therefore, certain miRNAs can be synthetically designed and generated to destroy functional or structural proteins of the virus. The ways for therapeutic treatments can be opened and used for patients who have previously been in contact with a vaccine or infection [39].
Antagomir is a novel type of engineered oligonucleotide and easy to be produced in the laboratory. It can specifically inhibit the function of the endogenous miRNAs. In this regard, antagomir can have a good effect on SARS-CoV-2 infected patients with severe conditions [40]. Therefore, they could be used effectively to regulate the whole cytokine storm to prevent the patient from having serious, life-threatening complications associated with the COVID-19. Especially, it can be used to maintain function of the key miRNAs that contribute to the inflammatory process in COVID-19 patients [41].
SARS-CoV-2 infections can be induced or inhibited by expression of certain miRNAs in respiratory cells that result in suppression of viral replication, pathogenesis, and anti-viral responses. MiRNA-based therapies have the main challenge of absence of an in vivo delivery method. Aerosolization of small RNAs with a micro sprayer is the most common and effective method for the delivery. Therefore, miRNAs can be delivered to the respiratory tract in respiratory infections as seen in COVID-19 disease [36].
Nanoparticles-based miRNAs could also be utilized as nano-vaccines to prevent SARS-CoV-2 infection. Potential miRNA containing nanoparticles can be developed as a nano-vaccines for SARS-CoV-2 infected patients. This nanoparticle containing nano-vaccines have several benefits over traditional vaccines. Their advantages are their specificity to infection site and minimal off-target effects. During the oral delivery, miRNAs nucleic acid structure may degrade in gastric environment. Therefore, new formulations of the miRNAs are investigated for oral delivery of the miRNAs. Chitosan, mannose modified chitosan, bovine milk-derived exosomes, bovine lactoferrin, lipidic, and PLGA-based (poly lactic-co-glycolic acid) nano formulations are utilized in the literature for efficient oral delivery of miRNAs [42-47]. Therefore, oral nano formulations of miRNAs are highly recommended for successful practice in clinics.
At present, there is no approved single specific antiviral therapy for COVID-19 and miRNAs have become a potential therapeutic tool. MiRNAs typically inhibit the translation and stability of host cell mRNAs; controlling genes involved in cellular processes. MicroRNAs have recently emerged as important regulators of viral infections. They might play an important role in the treatment of viral infections, as anti-viral and gene therapy in SARS-CoV-2 infection. The miRNAs of the host cell and SARS-CoV-2 are important to gain a comprehensive understanding of the virus and provide the basis for drug and vaccine development. Therefore, multifaceted targeting approaches are required in miRNA therapeutics for effectively combat SARS-CoV-2.
This work did not receive any particular grant from funding agencies in the commercial and public sectors.
We have no Conflict of Interest. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The figure of this article was created with BioRender.com.