Dear Editor,
Helicobacter pylori is a spiral, microaerophilic and motile bacterium originated by its polar flagella that is the etiological reason of chronic gastritis, peptic ulcer, gastric cancer, and extra-gastrointestinal diseases [1,2]. The human stomach is considered the only host of this bacterium. This bacterium has been residing in the human stomach for many years and has been co-evolved simultaneously with its host, especially since the horizontally transmitted condition is not observed in Helicobacter pylori. Therefore, several studies have shown that the human migration in the past can be noticed by studying the changes and evolution of this bacterium [3]. The multilocus sequence typing (MLST) is a reliable way to evaluate changes and the geographical relationship between different strains of the bacteria, which was first introduced in 2010. The sequence of seven housekeeping genes was studied in this method, and each isolate is considered a unique ST type that the distribution, origin, and evolution process of bacteria can be found by comparing the STs with the information recorded in the relevant databases [4].
In this study, we intended to compare the MLST data of Helicobacter pylori strains isolated from the Iranian patients with other regions and determine the changes and origins of the H. pylori strains of the said patients.
For this study, we first obtained the sequences of 7 housekeeping genes including atpA, efp, mutY, ppa, trpC, ureI and yphC belonging to 177 strains of Helicobacter pylori isolated from the Iranian population by searching the Popset database.
We then determined the sequence types using BioNumerics v5.10 databases (Applied-Maths, Sint Maartenslatem, Belgium), and the Iranian H. pylori strains data were supplemented with a number of data from hpEroupe, hpEAsia, hpAsia2, hspAmerind, hpMaori and hpAfrica1 from http://pubmlst.org/helicobacter. Finally, the phylogenic tree was constructed from http://pubmlst.org/helicobacter by UPGAMA (unweighted pair group mean mean). Moreover, the genetic diversity between the strains of the Iranians was investigated by phylogenic analysis and the minimum spanning tree calculations were evaluated in that respect [5].
Based on our phylogenic analysis, it was found that all the H. pylori strains isolated from the Iranian patients are in the hpEroupe clade (category) (Figure 1).
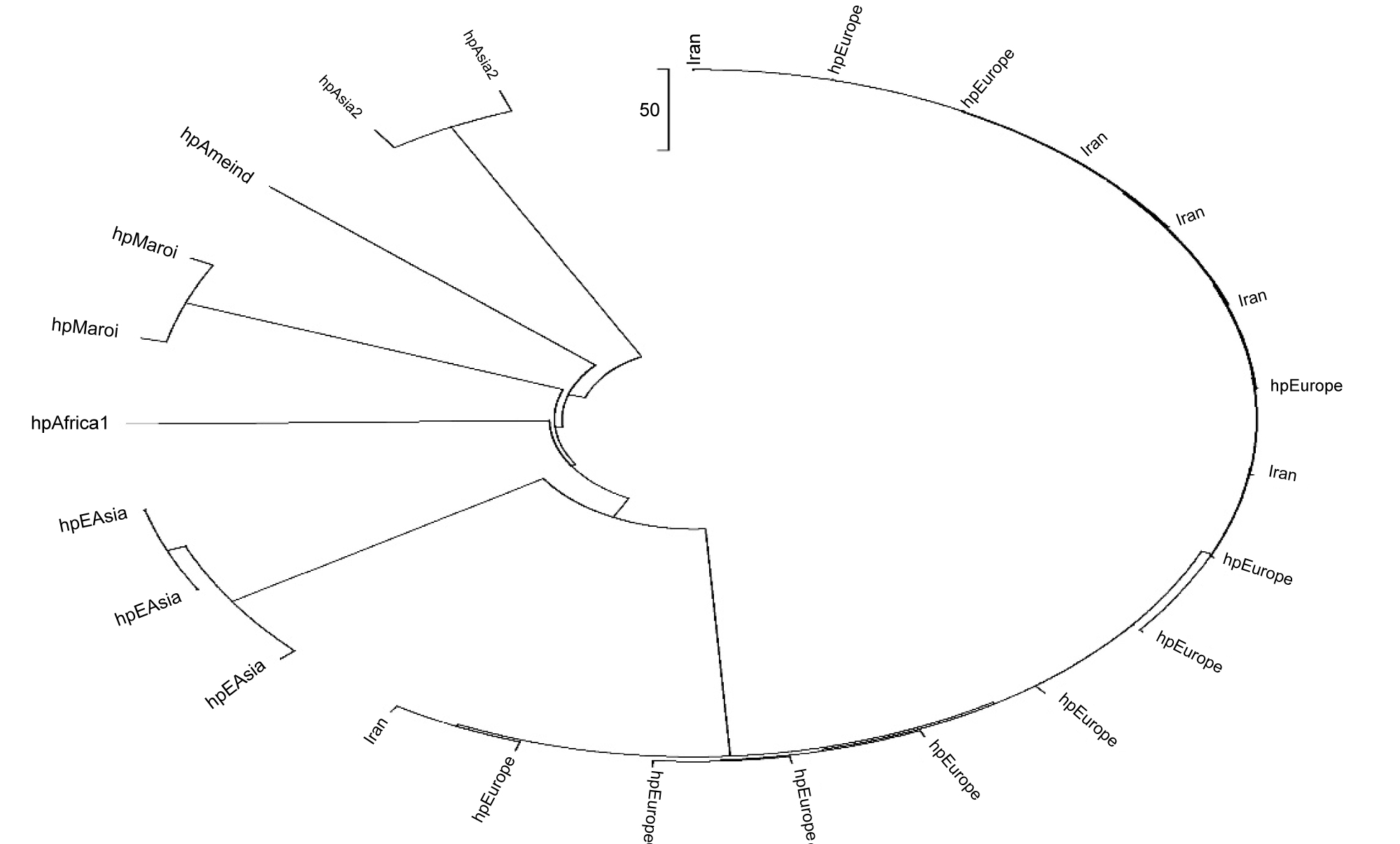 Figure 1: Population and phylogenetic structure of the H. pylori isolates.
View Figure 1
Figure 1: Population and phylogenetic structure of the H. pylori isolates.
View Figure 1
According to our results, it seems that the possible origin of H. pylori strains of the Iranian population belongs to the European population, especially from Spain, Germany, United Kingdom, Italy, and Greece. We also showed that the genetic diversity of the H. pylori strains of the Iranian population was significant. This phenomenon has a direct impact on the pathogenicity potential, the difference in cloning pattern and drug susceptibility, as well as the difference in results between Iran and Western countries (Figure 2). Furthermore, these changes may have occurred due to the compatibility of the strains with the Iranian population.
 Figure 2: Population structure of H. pylori strains. a) MLST based phylogenic relationship of H. pylori strains was isolated from Iranian patients by neighbor joining tree (Kimura 2-parameter); b) Minimum spanning tree of H. pylori strains reconstructed from MLST data.
View Figure 2
Figure 2: Population structure of H. pylori strains. a) MLST based phylogenic relationship of H. pylori strains was isolated from Iranian patients by neighbor joining tree (Kimura 2-parameter); b) Minimum spanning tree of H. pylori strains reconstructed from MLST data.
View Figure 2
In this study, we were looking for identifying the probable source of the H. pylori strains in the Iranian population. By evaluating all the MLST data on the Iranian Helicobacter pylori isolates, we demonstrated that the Iranian strains are in common clade with the Europeans and appear to be of hpEroupe root. The results of our study confirmed previous studies on the Iranian population [4,5]. Looking at the Iranian history and the examining the migrations of the Europeans during the past periods, the results were consistent with the results of our study (Figure 3).
 Figure 3: European migrations to Asia as proposed from H. pylori MLST data.
View Figure 3
Figure 3: European migrations to Asia as proposed from H. pylori MLST data.
View Figure 3
It seems that the European Helicobacter pylori strains were transferred to and replaced in Iran during Alexander's invasion, the Crusades, the World Wars, European immigration to Africa and the subsequent migration of Africans to the Middle East and South of India, the Muslim War and Victory, and the wars of Ottoman Empire in 1514 [4-7].
In summary, it seems that there is a historical connection between the human population and the circulation of Helicobacter pylori strains. Moreover, high genetic diversity among the Iranian Helicobacter pylori strains indicates that the strains of this geographical region are not clonal generation, and such changes may be due to compatibility with the host.