Introduction: Type 2 diabetes (T2DM) is a multifactorial disease with serious complications, particularly degenerative. Currently, several genes have been identified in its pathogenesis, including the 5,10-methylene tetrahydrofolate reductase (MTHFR) gene with an association of the c.677C>T and c.1298A>C variants. The aim of this study was first to investigate the frequency of these variants in Congolese subjects with T2DM and to compare them with healthy controls. Secondly, to establish a correlation between these genotypes and the degenerative complications observed in T2DM patients.
Material and methods: This was a prospective case-control study conducted over a two-year period. It involved 100 individuals with T2DM and 50 healthy, non-diabetic controls. The study method was based on DNA sequencing using the Sanger method, looking for the c.677CT and c.1298AC variants in the MTHFR gene.
Results: In the DT2 group, the study identified heterozygous genotypes MTHFR1298AC in 18% of T2DM cases and MTHFR 677CT in 17% of cases. The homozygous MTHFR 1298CC genotype was detected in 2% of individuals. It should be noted that combinations of these two mutations were identified in some T2DM subjects, namely: (677CT; 1298AC) in 1% of cases and (677CC; 1298CC) in 2% of cases.
In the control group, heterozygous genotypes MTHF R1298AC and MTHFR 677CT were identified in 6% and 4% of cases respectively. The difference was statistically significant (p = 0.024; p = 0.048).
Degenerative complications (stroke, neuropathy, ischaemic heart disease) were observed in 58.8% (20/34) of T2DM carrying the mutation exclusively in the heterozygous MTHFR 677CT form and in 44.1% (15/34) of T2DM carrying the mutation exclusively in the heterozygous MTHFR 1298AC form.
Conclusion: The c.677C>T and c.1298A>C variations of the MTHFR gene, identified in Congolese subjects with type 2 diabetes, could be predisposing factors for type 2 diabetes. A combination of these two variants would more often lead to degenerative complications.
MTHFR, c.677C>T and c.1298A>C variants, Type 2 diabetes, Congolese
Type 2 diabetes (T2DM), accounts for 80-90% of all diabetes [1].
It is a complex, multifactorial metabolic disease essentially favored by dietary imbalance and genetic predisposition [2]. The severity of its complications, especially cardiovascular, makes T2DM a major public health problem worldwide [1].
In 2021, the International Diabetes Federation (IDF) estimated the global prevalence of T2DM at 10% and it is expected to reach 15% (or 753 million people) in 2045 [3]. In Africa, it affects 22% of the population (or 24 million people) and is expected to reach 55 million by 2045 [3].
In the Congo, according to the national plan to combat non-communicable diseases (2013-2017), diabetes ranks second (10%) among chronic diseases after cardiovascular diseases [4].
Genetically, T2DM is a polygenic disease and there are several predisposing genes, including MTHFR (methylenetetrahydrofolate reductase). This gene, located at 1p36.3, contains 12 exons [5]. It is involved in homocysteine (Hcy) metabolism and several studies have shown that it is associated with T2DM [2,6].
The most commonly reported variants in the MTHFR gene are variant c.677C>T (p.Ala222Val) (rs1801133) on exon 4, and variant c.1298A>C (p.Glu429Ala) (rs1801131) on exon 7 [7-9]. These variants lead to the formation of a heat-labile isoform with reduced enzymatic activity (70% and 30%) for c.677C>T and c.1298A>C, respectively) [10], resulting in a relative deficiency in the remethylation process of Hcy [11] and finally leading to a high plasma Hcy concentration in addition to a drop in plasma folates for c.667C>T [7,9] whereas c.1298A>C is not necessarily associated with higher Hcy concentrations [8].
These variants are also associated with macrovascular complications (arterial hypertension, stroke and heart failure) or microvascular complications (retinopathy, nephropathy, neuropathy, diabetic foot and susceptibility to infections) [12].
In Congo, these variants have never been studied. Given the high prevalence of T2DM in the country and the frequency of these degenerative complications, we were interested in exploring these variants in this population. The objectives of this study were firstly to investigate these variants (allele and genotype frequencies) in Congolese subjects with T2DM compared to controls and, secondly, to establish a correlation between the genotypes identified and the degenerative complications presented by these T2DM subjects.
This was a prospective case-control study conducted over a two-year period (October 2020 to December 2022). Patients were recruited from the Department of Metabolic and Endocrine Diseases at Brazzaville University Hospital (Congo) and from the DI@BCARE diabetes management centre. Genetic analyses were performed at the Biochemistry and Molecular Biology Laboratory of the Tours University Hospital (France).
The study was conducted on one hundred patients with a clinical and paraclinical profile (blood glucose > 1.20 g/l) in favour of T2DM and fifty non-diabetic healthy controls, selected at random. The T2DM patients were divided into two subgroups: Thirty-four with macrovascular and microvascular complications and sixty-six without vascular complications.
Diabetic patients with pathologies (liver disease, hypothyroidism, cancer) that could influence Hcy concentration were not included in the study.
All consenting individuals assumed to be healthy were included as controls in this study.
Epidemiological investigation: Anthropometric parameters and sociodemographic data were collected on a pre-established form containing a range of data including age, sex and medical history.
Glycaemia testing: All selected patients were first tested for blood glucose using the hexokinase enzymatic method in serum (Abbot Glucose kit ref 3L82-22) [13]. It was performed at the Biochemistry Laboratory of the Brazzaville University Hospital.
Molecular analysis: After collection of venous blood on an EDTA tube, the samples were stored at -80 °C and then shipped to the Biochemistry and Molecular Biology Laboratory at Tours University Hospital (France).
DNA Extraction was carried out from leucocytes using the QIA symphony extractor from the Innovative Transversal Technology Unit of the CHRU Bretonneau Laboratories in Tours.
DNA concentration was measured using Nanodrop 2000 ® absorption spectrophotometry at 260 nm and DNA quality was assessed using the 260/280 nm ratio.
The technique used for genotyping the MTHFR gene was the PCR/Sanger method described by Smith [14].
The primers used were as follows:
F: 5'-CCTCTCCTGACTGTCATCCC-3'; R: 5'-GCCTTCACAAAGCGGAAGAA-3’ for the c.677C>T variant.
F:5'-TACCTGAAGAGCAAGTCCCC-3'; R:5'-ACAGGATGGGGAAGTCACAG-3' for the identification of the c.1298A>C variant.
Exon amplification was performed on an AB Applied Biosystems Veriti ® thermal cycler. PCR products were read after migration on a 1% agarose gel using the BIORAD Gel Doc™ XR+ Molecular Imager © .
The amplicons were then purified on a Macherey-Nagel™ plate to remove excess primers and nucleotides not incorporated during the PCR reaction.
Sequencing of the amplification products of the MTHFR gene was performed on the Biometra TRIO™ Analytikjena thermal cycler, using the Big Dye Terminator Cycle Sequencing kit with the same primers as for PCR. A purification phase was then performed on Millipore™ plate and then transferred to a MicroAmp™ plate to finally be placed in an Applied Biosystems™ 3500xL Genetic Analyzer DNA sequences.
The results were analysed using SeqScape Software v4 and Sequencing analysis Software v7 from Applied Biosystems™.
Statistical analyses were performed using Excel 2013 and SPSS ® 20.0 software. Data were expressed as percentages and allelic frequencies for qualitative data and as mean ± standard deviation for quantitative data. Comparisons were made using the Student's t test for quantitative variables and the χ 2 test for qualitative variables (such as allelic frequencies). Statistical tests were considered significant for a value of (p) < 0.05.
The mean age of the T2 DM subjects (Figure 1) was 52.2 ± 10.8 years, with extremes ranging from 30 to 83 years. Control cases had a mean age of 42.3 ± 7.1 years, with extremes ranging from 28 to 61 years. We found no significant difference between the ages of patients and controls (p = 0.184).
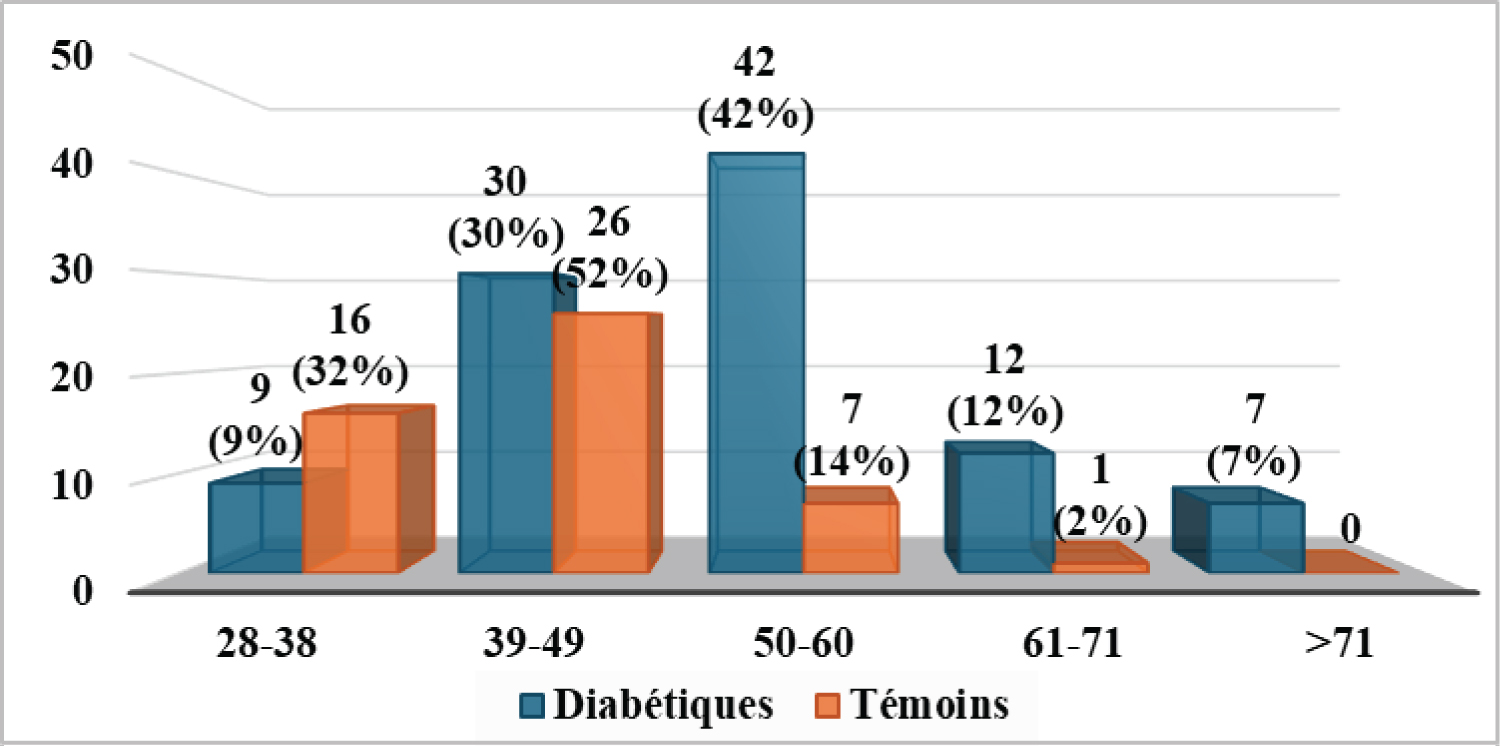 Figure 1: Age distribution for the two groups in the study population.
View Figure 1
Figure 1: Age distribution for the two groups in the study population.
View Figure 1
The gender distribution (Figure 2) for T2DM patients was 50/100 females, with a sex ratio of 1. In the controls, there was a clear predominance of females (36/50). There was a statistically significant difference between controls and controls with p = 0.01.
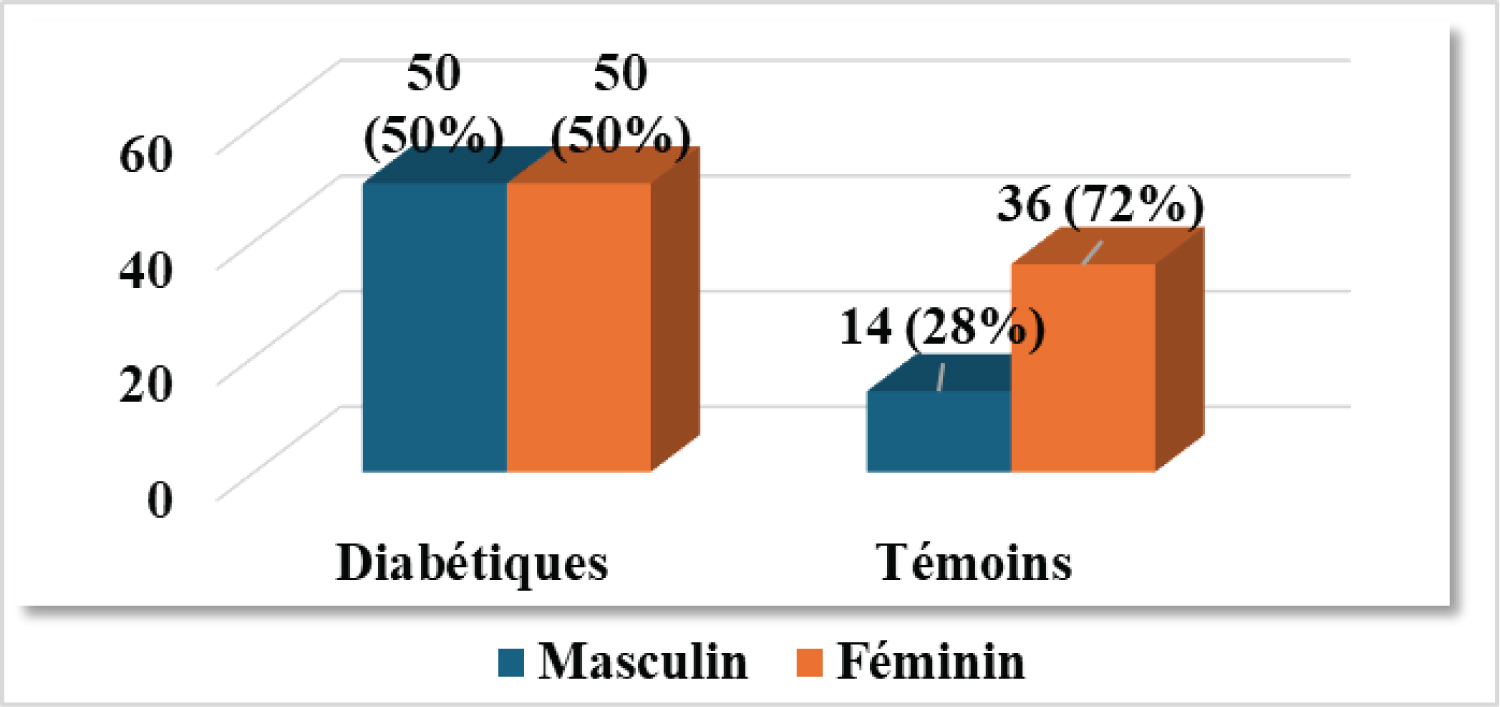 Figure 2: Gender breakdown of the two groups in the study population.
View Figure 2
Figure 2: Gender breakdown of the two groups in the study population.
View Figure 2
c.677C>T variant: 677C>T variation was found in 17% (17/100) of T2DM in heterozygous form only. It was found in 4% of controls (2/50). It should be noted that the homozygous 677TT genotype was not found in our series. The difference in frequency of genotypes identified was statistically significant between controls and T2DM (p = 0.024) with an increased risk of T2DM of odds ratio (OR) = 4.9 (95% CI: 1.1-22.2) (Table 1).
Table 1: Prevalence of C677T and A1298C mutations. View Table 1
Variant c.1298A>C: This variation was identified in both heterozygous and homozygous form. The heterozygous1298AC genotype was present in 18% (18/100) of T2DM and in 6% (3/50) of controls (Table 1). The homozygous 1298CC genotype was found only in T2DM (2%). As with the previous variation, the frequency of the latter genotypes was also significantly different (p = 0.024).
The A1298C variation was associated with T2DM at an OR risk of 3.99 (95% CI: 1.1-13.8).
Sequencing of the MTHFR gene also identified an association of mutated genotypes in T2DM: (677CT; 1298AC) in 1% (2/100) of cases; (677CC; 1298CC) in 2% of cases.
The other combinations identified in T2DM: (677CC; 1298AC) in 17% of cases and (677CT; 1298AA) in 16% of cases; p = 0.274 showing no difference.
Degenerative complications were observed in 34% (34/100) of T2DM patients. Of these patients, 58.8% (20/34) had the 677C>T variation in heterozygous form, while 41.2% (14/34) had the 1298AC variation. The latter was heterozygous in 93.3% (14/15) of cases and homozygous in 6.9% (1/15) (Table 2).
Table 2: Correlation of C677T and A1298C mutations with degenerative complications observed in T2DM. View Table 2
Numerous studies conducted in various countries on the c.677C>T and c.1298A>C mutations show a significant association with T2DM [2,5,15,16].
The results of our study revealed that the C677T mutation is more frequent in the heterozygous form in Congolese T2DM (17%) than in controls (4%) with a 95% CI [1.2-22.2], p-value 0.018. Our data are close to those reported in Egypt (18.3%) but significantly lower than those found in the Egyptian control group (18.3%) [16]. It should be noted that these results are not isolated; in fact, the same mutation has already been identified in Tunisia and Morocco in 31.4% and 34.9% of T2DM and in 29.4% and 46.5% of controls respectively [17,18]. In Brazil, 37% of T2DM and 43% of controls [19]. As observed in the literature, this heterozygosity is identifiable in both groups studied.
The homozygous 677TT form was not found in our study cohort. This is normal given the allele frequencies and the fact that the Hardy Weinberg equilibrium was respected in the T2DM group: C677T (chi 2 : 1.031; p = 0.597) and the Control group: C677T (chi 2 : 0.00; p = 1). However, it has been frequently reported in other studies. For example, in Egypt and Tunisia, it was observed in 20% and 27.5% of T2DM and 6.67% and 5.9% of controls respectively [16,17]. In Brazil, it is present in 9% of T2DM and 12% of controls respectively [19].
It should be noted that low values have been observed in the Afro-Brazilian and Afro-American diabetic community (1 to 2%) [20,21]. Furthermore, in African-Americans, the 677TT genotype has never been identified as in the present study [22].
These variations could be explained by the consequence of the presence of these variants: Hyperhomocysteinemia. In fact, these HHcy are related to ethnicity in that they are linked to diet, particularly folate deficiency [23,24].
Some studies have reported contradictory results, finding no link between the C677T mutation of the MTHFR gene and T2D [25,26].
Its allelic frequency was 20% in our series and is often reported in the literature [16,27,28]. Yan, et al., whose work was carried out on two types of populations, revealed that there is a predisposition to diabetes in Asian populations carrying this variant. However, in Caucasian populations, the authors found no link between the A1298C variant and diabetes [27].
Genotypically, the 1298AC heterozygosity was detected in our study in 18% of T2DM patients and in 6% of controls. Higher frequencies have also been reported in North African communities: In Egypt (50% in T2DM and 28.3% in controls) and Tunisia (42.5% in T2DM and 7% in controls) [16,28].
The homozygous 1298CC form found in our study in T2DM had low rates (2%). However, higher frequencies have been reported in Egypt (16.6% of T2DM and 8.33% of controls), Tunisia (42.5% of T2DM and 7% of controls) and Morocco (34% of T2DM) [16,18,28].
This low frequency can be explained by the allele frequencies and the fact that the Hardy Weinberg equilibrium was respected in the T2DM group (chi 2 : 4.35; p = 0.113) and controls (chi 2 : 0.985; p = 0.321).
Finally, our results, combined with those in the literature, suggest that the A1298C mutation predisposes to T2DM at high risk and appears to be more frequent in Caucasians.
There is some evidence in the literature that the combination of the C677T and A1298C mutations may act synergistically to reduce the activity of the MTHFR enzyme in T2DM by 70% and 30% respectively [8,29].
When this association is in the combined forms, they further alter the activity of the MTHFR enzyme.
In the present study, the forms detected were (677CC; 1298CC), (677CT; 1298AC) (677CC; 1298AC) and (677CT; 1298AA). However, the homozygous mutated form (677TT; 1298CC) was not found in this study.
These results are similar to those reported in North Africa (Egypt and Morocco), which have described the same combinations in T2DM, at varying frequencies [16,18].
Several hypotheses have been invoked: environment, lifestyle, ethnicity, genetic predisposition [28]. A future study focusing on an epigenetic analysis would be interesting, in order to investigate the existence of MTHFR gene-environment interactions.
Numerous studies correlate diabetic complications with MTHFR mutations [28,30,31]. Concerning the C677T mutation, it should be noted that some case-control studies show no association between C677T and the occurrence of degenerative complications of T2DM [25,31,32]. For the A1298C mutation, some studies have found no link between the mutation and predisposition to degenerative complications [18,31,33].
However, the present study significantly suggests that there is a relationship between degenerative complications and the mutations identified. In our study population, degenerative complications occurred in 34% of T2DM, in whom the mutations were detected either in heterozygous or homozygous form.
The C677T mutation was identified exclusively in heterozygous form in 58.8% of T2DM presenting with degenerative complications. In contrast, the 1298AC mutation was observed in 44.1% (15/34) of T2DM, of which 93.3% (14/15) were heterozygous. The homozygous form was observed in only 6.9% (1/15) of T2DM with degenerative complications.
The genotype combinations (677CC; 1298CC) and (677CT; 1298AC) were the most implicated in the occurrence of degenerative complications in Congolese T2DM.
We used an unpaired sample for the 2 groups.
Concerning gender: Data from the literature show that the C677T and A1298C polymorphisms associated with T2DM are responsible for the hyperhomocysteinemia detected more in male subjects [34,35].
The data from the present study show that, genotypically, the heterozygous forms (677CT; 1298AC) were mainly detected in male subjects (63.2%; 71.4%), while the single homozygous mutated form (1298CC) was found in both sexes (Table 2).
These results suggest that heterozygous forms (677CT; 1298AC) predispose mainly male Congolese subjects to T2DM.
Concerning age: The literature reports that the two mutations studied are mainly observed in young subjects [36].
However, our study reveals that both young and elderly subjects are affected. Indeed, the heterozygous forms (677CT; 1298AC) are observed in the 28-60 age group and the homozygous mutated form (1298CC) in a relatively older age group: 50-71 years. These results corroborate those of other published studies [18,37].
The study relied on a single method of analysis (sequencing of the MTHFR gene) and a small sample, and also the control group was unmatched.
The C677T and A1298C mutations in the MTHFR polymorphism gene are more frequent in type 2 diabetics (compared with control cases).
Their prevalence suggests that these mutations are risk factors predisposing Congolese subjects (especially males) to T2DM in the Congolese population.
In addition, genotype combinations (677CT; 1298AC) and (677CC; 1298CC) are more prone to degenerative complications.
Our thanks go to: French Embassy and Campus France for facilitating mobility; Laboratoire de Biochimie et Biologie Moléculaire of CHRU Bretonneau de Tours for genetic analyses; Formation Doctorale Santé Biologie Humaine de la Faculté des Sciences de la Santé de Brazzaville.
MHG, ME, and AC initiated the project. IMVR, AH, DC, VP, AC, carried out the experimentation. BE, MHG and WBE recruited the patients. IMVR, PH and BALM. AC reviewed the manuscript.
None.