Autism is still considered a very complex disease, since it has diverse etiology, with multiple factors apparently associated with its appearance. However, none of them reveals to be totally responsible for its development.
Nowadays, this disorder is considered to have a strong genetic component with the interaction of several genes. Furthermore, other diseases with well-known etiology might also be related with autism.
The aim of this review is to discuss, through bibliographic search, genetic factors involved in autism development.
Some of the candidate genes of idiopathic autism (90-95% of all cases) related to brain metabolism are AVPR1a, DISC1, DYX1C1, ITGB3, SLC6A4, RELN, RPL10 and SHANK3.
All these genes are responsible for symptoms that can explain the origin of this condition. Autism is a multifactorial disorder in which genetic and environmental factors interact, triggering its development.
Autism spectrum disturbances, Genes determining autism, Idiopathic autism
Since the beginning of human existence, communities exclude or neglect those that deviate from normal standards at physical or mental levels. A lot comes from the unfounded myths and believes and from the unknown itself. Nowadays, interest on this population is increasing and consequently research is focusing on deepening knowledge on these disturbances aiming their integration on a society build apart from those who have special needs.
The word "autism" comes from "autos", greek word that means "itself" or "to itself". It is a syndrome with extremely variable phenotype, being considered a disturbance of human development.
Reports on the increase in autism prevalence in the population, studies concerning treatment efficacy and high costs in education and care of these patients has made this pathology a focus of great interest in public health.
It is estimated that the prevalence of Disturbances of Autism Spectrum (DASs) is 0.5-1% and seems to increase 15% each year. The discussion between a real increase of these disorders or a possible improvement in diagnosis, remains [1].
The study of autism started 7 decades ago. It was described for the first time in 1943 by the Austrian doctor Leo Kanner, in his scientific paper "Autism Disturbances of Affective Contact" [2]. In this work, the author started to group a set of apparently characteristic behaviours, present in eleven children followed by him, mainly boys. In theory, these behaviours could help to identify children with this disturbance, that showed perceived, emotional and cognitive deficit [2-4]. In the two following decades and despite the publication of that study, there was a lack of interest on this disorder.
However, first results pointed to the similarity between the terms Autism and Child Schizophrenia, that probably led to their incorrect differentiation. Confirming this fact, the concept of autism had already been introduced by Bleuler in 1911, by referring to schizophrenia as a limitation in human relationships and the outside world [5]. In 1971, the two concepts were finally dissociated due to differences in symptoms (such as the age of onset of first symptoms), family history and differences in response to treatment of adults suspected to have schizophrenia/child autism [1].
After Kanner, Hans Asperger [6] published a paper entitled "Childhood Autistic Psychopathology", based on the study of four children described as carriers of the same resistance in the establishment of social contact, and the same pattern of restrict interests, but having sophisticated language skills [6]. At this moment, the term Asperger Disturbance appeared [4,7]. Nowadays, it is easily distinguished from classic autism [8].
After the work of these two scientists, other clinical studies followed up, however, only with the publication of the 4th edition of the Diagnostic and Statistical manual of Mental Disturbances (DSM-IV), of the American Association of Psychiatry, autism was classified as a type of Global Disturbance of Development (GDD) [4,9]. Nowadays, it is described as a change in normal development of a child that shows abnormal "communication, socialization and restrict and stereotyped behaviours" [4]. Among GDDs, it is possible to distinguish different types: Asperger syndrome; Global Disturbance of non-specified Development (GDD-NS); Rett Syndrome and Child Disintegrative Disturbance [4,5,9], (Table 1).
Table 1: Diagnosed types of GDDs according to the classification of DSM-IV-TR [5]. View Table 1
According with Lorna Wing, the degree of autism varies with the deficits observed over time at social interaction, verbal and non-verbal communication and the use of imagination, being called the "Triade of Social Impediments" [10]. Deviations of these disturbances result from their very heterogeneous and mostly unknown aetiology [11]. So, description of autistic spectrum goes far from strict definitions, including individuals with low cognitive potential that reveal deficit on reciprocal verbal communication and repetitive behaviours together with mental retardation and those having high cognitive potential that even having deficit in social interaction with repetitive and stereotyped behaviours, do not show retardation in speech development (like in the Asperger syndrome) [4].
GDDs (among them are ASDs) involve a group of clinical changes with early onset where diverse basic areas of behaviour and development are simultaneously compromised. Rett syndrome and Disintegrative Disturbance of Second Childhood are also included in this group [5,12].
Although now a days knowledge concerning autism is much more prefound, it still surprises due to the diversity of characteristics that patients can show.
Usually, the autistic child has normal physical [3,13]. However, these children also show an irregular profile of development that is detectable in the first three years of life, being present till adulthood. The Triade of Social Impediments is characterized by a strict and continuous pattern with intelligence levels varying from mental retardation to an extraordinary performance in certain cognitive domains (like music, arts, mathematic, memory) or savant capacities [3,4,10,14]. Although 80% of autistic children show mental retardation, savant capacities can exist, however the global intelligence ratio is low [3,12,14]. The difference between mental retardation and autism should be pointed out: The first one shows a uniform development deficit; while the last presents an irregular profile, with differentiated degrees of commitment.
The following classification and diagnosis systems allow to distinguish autism from other disorders: International classification of Diseases of the World Health Organization and the Manual of Diagnosis and Statistics of Mental Disorders from the American Academy of Psychiatry (DSM-IV). In these systems the term Child Autism was replaced by Autistic Disturbance, officially separating it from Asperger syndrome [15].
According with Siegel [8] echolalia (child repeats the same sound consecutively), incapacity to play symbolic games, prenominal inversion (use of the third person instead of the first), abnormal voice sound, absence or deficit of speech, peculiar relation within animate objects, stereotyped interests, repetitive behaviours, deficit in social, emotional and interests share and trouble in establishing visual and physical contact are common characteristics of autism [8]. Other symptoms frequently observed in these individuals are hyperactivity, attention deficit, impulsivity, aggressivity and self-aggressivity [13].
Individuals with Asperger disturbance reveal social deficits and restricted interests. However, their speech capacities remain intact, or show speech capacities above average for their age [4,8].
Individuals with Global Disturbance of non-specified Development show changes in one or more developmental areas, however, these are not enough to be included in the autistic or Asperger disturbances, since they are lower in number or intensity [16].
The phenotypic heterogeneity or high variability observed in behavioural standards and in social and communication capacity gave rise to the predominant use of the term Autistic Spectrum Disturbances [15].
The distinction between autistic disturbance and the remaining ASDs is more complicated due to the simultaneous occurrence of one or more psychiatric and neurological disorders in most patients (72%), including: Attention/hyperactivity deficit, tic disorder, dyspraxia (neurological motor dysfunction), dyslexia (reading, writing and spelling difficulty), obsessive-compulsive disorder, phobias, anxiety, humour, sleep and feeding disturbances [17].
In the Portuguese population, the prevalence of DASs is one in a thousand children [3,17]. It is estimated that 60-70 in 10,000 individuals suffer from a Global Disturbance of Development, and 20 in each 10,000 are disturbances of the autistic spectrum (one third being Asperger disturbance). These values make DASs the most frequent forms of child developmental disturbances, and a rise is being observed over the last decades [18]. However, prevalence differs between gender, being four times higher in men. Even so, an autistic female is usually more affected at cognitive level [3,17].
Since Kanner studies, some characteristic aspects of autism started to be analysed in the personality of parents and affected children. Initially, the cause of autism was associated with parents emotionally and socially cold. Later, scientific studies associated autism to mental retardation, genetic syndromes, epilepsy and other neurological conditions that had the organic bases of autism and related disorders [5]. These have strongly contributed to elucidate the biological aspects of ASDs and demonstrated that these are complex conditions, with multiple aetiologies and diverse degrees of severity. Although evidences demonstrate that most cases of ASDs result from genetic causes, the expression of those genes can be modulated by environmental factors from the gestational period. It should be emphasized that several deficiencies with cerebral involvement associated with ASDs, occur during the first and second trimester of pregnancy (teratogenic effect) [1].
Although a large study using microarray tests performed by Shen, et al. [19] observed that 80% of analysed autistic children had a normal genome, several other studies point for the existence of a narrow correlation between genetic predisposition (above 90%) and environmental factors [4]. Some consistent changes during gestation of affected individuals are known, however, pre-natal diagnosis is still not possible, mainly because no physical traits can be identified. The first alert sign is the deficient social interaction of the child, mainly observed between 6 months and 3 years of age [15].
In autism aetiology, "it is observed a drastic decrease in risk for family members of the second or third degree, pointing to the involvement of multiple interactive genes", that influence each other [20]. However, although numerous investigations at the pharmacological, pathological, electrophysiological and imaging levels have been done, ASDs aetiology is still under discussion [21].
Since it is still not possible the use of genetic markers for this disorder, diagnosis is usually based on meticulous investigation of the history of the patient and family inquiry (cognitive and behaviour capacities) according with Autism Diagnostic Interview-Revised (ADI-R) and the Autism Diagnostic Observation Schedule (ADOS) [16].
It is believed that autism appearance is related with some brain anomaly, not yet defined, associated with a genetic cause. Since ASDs show a very heterogenous aetiology, no definitive medical test or cure for this type of disorders has been developed yet. For this reason, only in very few cases definitive diagnosis happens before twenty-four months of age (a well established diagnosis is normally done between 3 and 6 years of age) [22].
In this way, evaluation should be done by one or more professionals with medical background and several years of clinical experience. The doctor tries to investigate, through several exams, the existence of disorders with known causes that can lead to a child autistic picture, such as the X-fragile syndrome or the Rett disturbance [15].
Clinical genetic tests can also be performed, and the usually used strategy involves, in the first place, a chromosome microarray analysis that allows the detection of chromosome copy number variation as well as the existence of chromosomal deletions or duplications of considerable size. The second approach involves molecular DNA tests for specific genes or even whole genome sequencing. For instance, patients can be tested for Fragile X syndrome by the analysis of specific single gene or, in the presence of a specific feature/condition, search for a set of genes that have been associated with those characteristics.
Autism genesis is divided in Secondary Autism (with environmental agents, chromosomal anomalies or identified genetic changes) and in Idiopathic (without known cause and involving more than ten genes), being this last one the most common [3].
Several genetic diseases are related with an increased risk of ASDs, being responsible for 5-10% of total diagnosed cases. Since autism prevalence is higher in men, several studies have proposed that the genetic influence of the X chromosome increases the susceptibility for ASDs [14].
Patients usually show developmental problems, mainly speech and motor retardation, or even, autistic behaviours. Since the locus of the main involved gene is the X chromosome (Xq27.3), the probability of X-fragile syndrome development is increased in males. In most cases (99%), the disease is related with the loss of function of the FMR1 gene, that results from the presence of multiple copies of the triplet repeat CGG in the 5' untranslated region (5' UTR) of that gene. The FMR1 protein (FMR1P - fragile X mental retardation 1 protein) is the product of the FMR1 gene, and is mainly present in neuron cytoplasm, being involved in important processes like ribosomal translation and maturation of synapse structure and function, having a suppressor effect at the post-synaptic region [14,23].
The number of CGG repeats is variable and can be classified as a pre-mutation (59-200 repetitions) or full mutation (above 200 repeats). In the presence of a pre-mutation transmitted by the mother, the meiotic instability might result in a complete mutation [14].
Autism has a high prevalence in XFS individuals, reaching 25-33%. The FMR1 protein can affect the expression of several genes involved in autism development. However, XFS is distinguishable from autism by the presence of a specific biomarker, the abnormal expansion of the CGG repeat in the X chromosome [14,23].
Down syndrome is the most frequent cytogenetic anomaly in patients with ASDs (7-10% of individuals are carriers of ASDs), followed by the one found in the region 15q11-13 (present in 1-4% of autistic patients). When the 15q11-13 region suffers duplication or inversion, a high incidence of epilepsy in childhood is observed, as well as muscular hypotony and motor coordination problems, together with strong or moderate mental retardation, absence or delayed speech and severe hyperactivity. If deletions occur in the referred chromosomic regions, maternal or paternal, imprinting diseases will develop: Angelman and Prader-Willi syndromes respectively [24].
Rett syndrome associated with the MeCP2 gene, is a dominant disease associated with the X chromosome, being considered one of the factors responsible for a small number of autism cases. This syndrome is usually lethal in boys. The most frequent mutations occur in exons 2, 3 and 4 of the mentioned gene [14].
Some researchers refer that epilepsy episodes in individuals diagnosed with ASDs, occur in a casual form. Data show an increased risk of epilepsy in ASDs individuals, resulting from a Central Nervous System anomaly characterized by neuropathological changes. One third of autistic individuals develop epilepsy, since dysfunctions in the CNS strongly increase the risk of developing it. The prevalence of autistic children with epilepsy varies between 5 and 46% [13,21].
This condition can be associated with exposure to infections before birth and subsequent inflammatory response. In the same way, autism can result from this phenomenon since it shares with schizophrenia several brain morphological changes [1].
According with Meyer et al. [1] infections in women during the critical periods of pregnancy can lead to acute inflammation associated with the presence of cytokinin in the foetal system, particularly at brain level. This affects negatively neuronal development processes: Proliferation, migration and cell survival. In this way, acute inflammation during foetal neuronal development can lead to the formation of abnormal neuronal substrates that interfere with social, cognitive and emotional behaviour processes, among others.
The AGTR2 gene, present in the X chromosome (Xq22-q23), seems to be involved in the development of mental retardation in autistic individuals, since these patients show deletion of this gene. However, the Xq13-q21 region that contains genes of the neuroligins family is even more important in the development of this condition. Neuroligins are crucial in processes of cellular interaction, through adhesion molecules, between neuronal cells [24,25].
Approximately 75-80% of autistic children suffer a severe mental retardation [5].
Idiopathic autism is the type of autism with unknown aetiology, representing 90-95% of total diagnosed cases. Several genes have been tested as possible candidates for its development, and, so far, more than 10 genes have been identified [5,11].
Several psychiatric diseases have strong evidences of genetic involvement in their origin, and among them are schizophrenia, bipolar disturbance and autism. In 1977, a study with mono and dizygotic twins described for the first time the genetic predisposition of autism [26,27].
Nowadays, population studies suggest that the model that better describes DASs is multifactorial [13,20,28,29] with a concordance of 60-92% in monozygotic twins and 0-10% in dizygotic twins [30]. Differences found in studies between monozygotic twins support the multifactorial model, demonstrating the importance of environmental factors.
Several studies were performed to clarify genetic factors associated with the disease. Autism symptoms that suggest a strong genetic component are convulsions, mental deficiency, neurons and synapse decrease in amygdala, hippocampus and cerebellum, increased size of encephalon, and increased level of circulating serotonin. Even studies with monozygotic twins show a significant concordance, as opposed to dizygotic twins. Non-twin siblings present a risk of developing autism ranging from 0-30%, and this risk is much higher than in the general population [31].
The comparison of the mentioned populational groups, as well as the difference between men and women, shows epistatic effects (hierarchy between alleles) that involve an interaction between several genes, suggesting the role of environmental factors [32].
Studies on monozygotic and dizygotic twins, adopted children and their families helped the establishment of a strong genetic component in autism. Still, analysis performed on the genome of autists failed in the establishment of consistent signs of linkage. None of cognitive and affective diseases, as well as certain psychoses, follow a heritage pattern according with Mendel's laws, reinforcing the hypothesis of the involvement of multiple genes [24].
Since it is a complex disorder with the involvement of several variants, each contributing with a reduced risk to phenotype, the identification of susceptibility genes has been difficult. Moreover, hypotheses that support the presence of synergism and/or epistas is between multiple candidate genes are based on chromosomic changes that are not always enough for disease development [24]. So, traditional methods of linkage used in most studies, are insufficient to detect modest genetic effects, in typically small samples. It is then necessary to select candidate genes based on the type of coded proteins that play an important function in autism development [33].
A big number of involved chromosomes suffer translocations and inversions. These changes result in interruption of genes as well as deletions and duplications that are responsible for differences in gene expression. The most frequently reported mutations are duplications in 15q11-q13 and deletions in 2q37, although most of them are undetectable by karyotype analysis [34].
According with Freitag [27] some chromosomic deletions in 2q37, 7q31, 22q11 and 22q13.3, are also important in the cytogenetic evaluation of autism. Chromosomic micro deletions responsible for certain syndromes, are associated with the appearance of secondary autism: Velocardiofacial syndrome, DiGeorge syndrome and Facial Conotruncal anomaly syndrome.
Several genes have been associated with the development of autism. However, the appearance of this condition seems to involve the combination of changes in several genes, behaving in an additive manner.
In this study, brain-related genes though to have a role on the prevalence of this disorder are described (Figure 1). However, many other genes seem to play a role in the development of this condition, as is the case of genes encoding ion-channels.
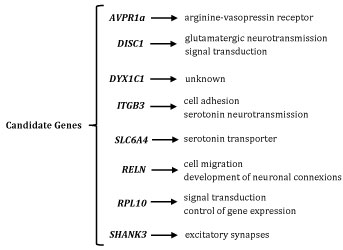 Figure 1: Brain-related genes though to have a role on the prevalence of Autism. View Figure 1
Figure 1: Brain-related genes though to have a role on the prevalence of Autism. View Figure 1
This gene is located at the long arm of chromosome 12 and encodes the arginine-vasopressin receptor (AVPR). Mutations in this gene lead to changes in the number of these receptors. The first report on changes in this gene associated with ASDs dates from 2002. In this study, researchers genotyped two microsatellite polymorphisms from the 5' flanking region of the AVPR1a gene. This work found significant transmission disequilibrium between autism and one of the microsatellite markers. Moreover, ten single nucleotide polymorphisms were identified by screening 2 kb of the 5' and coding region of the gene [35].
The CNS shows differences in receptor distribution that might interfere with the effects of the AVP system, among which are: Vocal position; sexual and parental behaviour; aggressivity; and social recognition. The AVP system reveals stronger effects in males [28].
According with Wassink et al. [28] autism susceptibility can increase by changes in the gene coding for the receptor 1a of the AVP system (AVPR1a). These changes lead to deficient social interaction, lack of parental behaviour, severe aggressivity, among other symptoms.
This gene is present in the locus 1q42 that contains the TRAX and DISC2 genes (involved in the regulation of DISC1 gene expression). The DISC1 gene codes for a protein with an essential function in growth and neuronal migration, in synaptogenesis, in glutamatergic neurotransmission (whose molecular processes of synaptic modification are among the most common in all CNS), and in cAMP signal transduction [29].
Kilpinen, et al. [29] were able to establish an association between autism and a DISC1 intragenic microsatellite (D1S2709). A single nucleotide polymorphism in the DISC1 gene (rs1322784) was also associated with male patients having ASDs [29].
The discovery of interaction between the TRAX and DISC2 genes and the DISC1 gene raised a new hypothesis that is based on the possibility of molecular mechanisms leading to brain dysfunctions at the affective and cognitive levels.
It is present in locus 15q21 and codes for a protein whose role is still not completely understood. However, it has already been established that it is expressed in a subgroup of human neuronal and glial cells, being the first candidate gene for dyslexia and the second associated with language disturbances. In the presence of genetic change, like a translocation, the gene is interrupted and can lead to two variants. One of these is related with the loss of attachment place to several linking factors. The second leads to the appearance of a premature stop codon and consequent loss of four amino acids at the end of the coded protein [25].
Dyslexia is a disturbance characterized by problems in learning to read or write (independent of the intelligence degree, social extract or interests of the individual). This phenotype has some interest in the study of ASDs since it might represent other disturbances with language development delay. However, it doesn't seem likely to be a very relevant gene in autism development [25].
It is located in chromosome 17 (q21.3) and codes for β-integrin 3. This protein is present in cell surface, especially in platelets. Integrins are known by their participation in cell adhesion and in metabolism and neurotransmission of serotonin (5-HT). Integrin receptors have shown an important role in signalling, being able to influence transcription and translation [30,36].
According with Napolioni et al. [30] the presence of a single nucleotide polymorphism located at the 5' terminal of theITGB3 gene, is strongly related with high plasma levels of 5-HT, one of the characteristics most frequently found in autists (around 30% of autistic individuals) [36,37]. Allelic variants of theITGB3gene were identified in some studies, both isolated or interacting with allelic variants of the gene coding for 5-HT transporter (SLC6A4 gene, located close to the ITGB3 gene) [30,36].
Moreover, Schuch, et al. [38] analysed five single nucleotide polymorphisms in the ITGB3 gene. These researchers observed small association of the rs15908 and rs12603582 polymorphisms with symptoms of echolalia, epilepsy and aggressivity, characteristics of ASDs [38].
Genes that influence the serotonin system are strong susceptibility genes, since selective drugs for the serotonin system reveal more efficacy in the treatment of some misbehaviour found in autism (like disturbances related with anxiety and aggression) [36].
This gene is in the long arm of chromosome 17 and codes for the serotonin transporter. This is a chemical substance that acts at brain level, through signal transmission between neurons (synapses). The role of serotonin system in autism is not totally clarified yet, however, it is known that polymorphisms of this gene can modulate the recapture of the transporter, explaining the occurrence of hyper-serotonin in some autistic individuals [36,39].
According with Longo [5] insertion/deletion polymorphisms located at the promoter of the SLC6A4 gene have been identified. Serotonin levels in synapses are regulated by its transporter and, when this one becomes active, serotonin builds-up in blood platelets instead of rising in synapses (where it is low). This can lead to important behaviour changes that determine the appearance of autism [5,36].
Adamsen et al. [40] identified three heterozygous mutations in the SLC29A4 gene: c.86A > G (p.Asp29Gly) in two patients, c.412G > A (p.Ala138Thr) in five patients and c.978 T > G (p.Asp326Glu) in one patient. Expression of mutations p.Ala138Thr and p.Asp326Glu in cells revealed significant reduced transport uptake of serotonin and dopamine, linking these mutations to ASD although with low brain serotonin. Dysfunction of this protein is speculated to raise serotonin prenatally, exerting a negative feedback inhibition through serotonin receptors on development of serotonin networks and local serotonin synthesis [40].
This gene is present in locus 7q22 and codes for a protein with an important role in migration of diverse cell types and in the development of neuronal connexions. It is believed that the RELN gene can affect the development of autism [33].
According with Skaar et al. [33] one of the most important effects of the deletion of this gene in rats, is the abnormal formation of cerebral cortex. Moreover, the study of this gene revealed its importance in the correct formation of brain structures due to the orientation that they confer in migration of neuronal precursors. According with these data, the RELN gene is related with several neurogenetic diseases (like schizophrenia and bipolar disorder), that show low levels of mRNA and RELN protein in multiple brain areas. The following genetic changes can be pointed out: Expansion of a trinucleotide polymorphic repeat (GGC) and haplotypes with replacement of two bases of one exon [24,33].
Moreover, Tian [41] identified two mutations in the RELN gene, and the g.504742G > A polymorphism, present in exon60, showed a significant association with autism in a Chinese Han population.
Some studies reveal linkage picks in the long arm of chromosome 7 that contains the RELN gene, and other susceptibility genes for autism, namely in the region 7q22-q33 [24,33]. Attention should be given to this gene, since changes observed in cerebellar neurons have impact in autism development [24].
It is present in chromosome Xq28 and codes for a family of ribosomal L10 proteins, that participate in the control of gene expression and are probably involved in cell signalling pathways. Mutations in the RPL10 gene (involving amino acid replacement) allow translation but a premature stop of protein synthesis occurs [42].
There is a marked difference in autism prevalence between males and females, being men the most frequently affected, suggesting a possible involvement of the X chromosome. The existence of linkage with markers in Xq22.3, Xq13-q26 and Xq27-q28 has been demonstrated. Changes in genes present in the region Xq28 lead to predisposal to mental retardation, that is frequently associated with autism [42].
Klauck et al. [42] have identified two missense mutations in the RPL10 gene in two independent families with autism. These mutations are the amino-acid substitutions L206M and H213Q at the C-terminal end of RPL10 protein that leads to changes in the regulation of the translation process. These researchers suggested as a model for the development of autistic disorder, that a change in translational function has impact on cognitive functions mediated through the limbic system [42].
It is present in chromosome 22 (q13.3) and codes for a protein (expressed at cortex, hippo campus and cerebellum levels) involved in excitatory synapses in front of the pre-synaptic active zone, and in the formation of intracellular protein complexes, by the establishment of connexions that favour protein interactions [43].
Several studies refer the identification of duplications in the SHANK3 gene that seem to have an important role in autism [43]. Cases of deletion of 22q13.3 with interruption or removal of the SHANK3 gene have been described. The resultant phenotype includes delay in expressive language, severe mental retardation and autism [43].
A single nucleotide polymorphism (rs9616915) is a T > C change in exon 6, that leads to a substitution of isoleucine to threonine and directly affects SHANK3 gene function. Mashavekhi et al. [44] were able to associate this mutation to increased risk of autism development.
Several advances in the study of autism genetics have been done, leading to drastic changes in the concept of autism, in the last decades.
According with the complexity of disturbances of the autistic spectrum, the analysis of isolated genes is not conclusive. It is very unlikely that any study performed with a candidate gene, covers a high percentage of sample population, consequently its inclusion in phenotype is difficult to identify.
Nowadays, most genetic studies focus on specific regions of chromosomes according with the probability of the corresponding genes being related with the observed characteristics of the disease. Obtained knowledge on genetic or environmental factors is very important for correct diagnosis and treatment and may allow prevention of the disorder.
A lot is still to be clarified in the autism spectrum disturbances, since it is likely that several susceptibility factors are involved and some still to be discovered. It is urgent to identify genetic variants not yet detected by the conventional genotyping studies, focusing on simultaneous analysis of multiple loci.
Diagnosis criteria used for autistic spectrum disturbances, have been improved. The ideal diagnosis would be the identification of genetic variants responsible for specific autistic phenotypes.
It is undeniable the contribution of heritance in autism disease but without considering environmental factors, it becomes insufficient to explain this complex disease.
Authors have equally contributed to the elaboration of this paper.
No funding was used in this study.
Authors declare that there are no conflicts of interest.
This article does not contain any studies with human participants or animals performed by any of the authors.