Journal of Genetics and Genome Research
Application of Synthetic Standard Curves for Absolute Quantification of Hepatitis A and E by Real-Time PCR
Renata Santos Tourinho1, Camilla Rodrigues de Almeida1, Andreza Salvio Lemos1, Noemi Rovaris Gardinali1, Yasmine Rangel Vieira1, Jonas Schmidt-Chanasit2 and Vanessa Salete de Paula1*
1Laboratório de Desenvolvimento Tecnológico em Virologia, Instituto Oswaldo Cruz - FIOCRUZ, Brazil
2Bernhard-Nocht-Institutfür Tropenmedizin-BNI, Germany
*Corresponding author:
Vanessa S de Paula, Pavilhão Helio e Peggy Pereira, sala B219, Instituto Oswaldo Cruz - FIOCRUZ, Cx Postal 926, Av. Brasil 4365, CEP: 21045-900, Rio de Janeiro, RJ, Brazil, Tel: 5521-2562-1876, Fax: 5521-2270-6397, E-mail: vdepaula@ioc.fiocruz.br
J Genet Genome Res, JGGR-2-013, (Volume 2, Issue 1), Short Communication; ISSN: 2378-3648
Received: February 26, 2015 | Accepted: March 20, 2015 | Published: March 23, 2015
Citation: Tourinho RS, de Almeida CR, Lemos AS, Gardinali NR, Vieira YR, et al. (2015) Application of Synthetic Standard Curves for Absolute Quantification of Hepatitis A and E by Real-Time PCR. J Genet Genome Res 2:013. 10.23937/2378-3648/1410013
Copyright: © 2015 Tourinho RS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Short Communication
Hepatitis A, which presents a major health problem globally, is caused by hepatitis A virus (HAV). This virus, whose primary site of replication is the liver, is the most common agent causing acute liver disease worldwide [1]. Hepatitis E virus (HEV) is the most recently discovered of the hepatotropic viruses, its genome having been identified in 1991. Despite the similarity with hepatitis A virus in their pathogenesis and transmission form, HEV has a zoonotic potential [2].
Two major challenges of virus analysis include a lack of adequate information in infectivity and the difficulty to cultivate these viruses in vitro. Conservatively, immunological, nucleic acid-based methods, and cell culture have been applied as molecular techniques for virus analysis [3,4]. The disadvantages of these methods have incited for new detection approaches that are rapid, sensitive and specific as the real-time PCR technique. Real-time viral detection provides sufficient information regarding multiple steps in infection process at molecular level, which is valuable for the prevention and control of viral infections [5].
Conventionally, in absolute quantitative real-time PCR detection, the viral load is measured as copy number per cell or per total RNA/DNA following a transformation of the data using a standard curve. A standard curve is generated by performing a real-time PCR using a dilution series of template DNA made from plasmid DNA or, more recently, DNA oligonocleotides. The current literature suggests that there are advantages and disadvantages of using different templates for the construction of a standard curve in the absolute quantification [6]. As the results of our laboratory cost-benefits studies, we could observe that, despite displaying reliable results, using plasmid DNA as template is more expensive and takes longer to synthetize than DNA oligonocleotides. The advantage of using DNA oligonucleotides is that it can be custom synthesized, and only requires the nucleotide sequence information.
Over the past decade, synthetic biology has taken advantage of a comprehensive inventory of biomolecular parts [7] and it is providing alternative diagnostic tools for viral detection and monitoring. This report highlights the application of synthetics standard curves for quantification of hepatitis A and E genomes by real-time PCR. The aim of this study was to examine the potential of two DNA oligos for hepatitis A (HAV) and hepatitis E virus (HEV) genomes quantification that could be adopted readily into established testing protocols.
The HPLC-pure oligonucleotides specifying the 89bp HAV (5' non-coding region) and 74bp HEV (ORF3 region) amplicons were custom synthesized by IDT® (CoralVille, USA) (Table 1), corresponding to the same genome fragments used for plasmid DNA standard curve construction. Following manufacturer's specifications, the DNA oligo was dissolved in DNase/ RNase-free distilled water to 100pmol/uL which is equivalent, approximately, to 1013 DNA molecules/uL according to the Avogadro number calculation - ([DNAg/uL]/ Plasmid size (bp) x 660) x (6,022x1023). The standard curves were generated by performing a TaqMan real time PCR, previously described by De Paula et al. and by Jothikumar et al. for HEV [8,9]. In this study, the plasmid standard curve was replaced to ultramer template ten-fold dilution series (10-1-10-20). The specific primers and probes were used according to the literature above. After determining the dilution range for each synthetic curve, they were assayed along with plasmid DNA as standard curve to compare their quantification in copies/uL. Therewith, the absolute quantification using DNA oligo as standard curve could be performed using a panel of 14 HAV and HEV available samples with known quantification.
![]()
Table 1: Hepatitis A and E synthetic standard curve sequences
View Table 1
Firstly, in our experiments, we could observe that the range dilution for HAV synthetic curve was 106-100 molecules/uL and for HEV 106-101 molecules/uL (Figure 1). The difference between the dilution ranges of both oligo DNAs could be explained by HEV genome quantification results. It was detected non-specific amplification at high dilutions or at a low copy number. The Ct value of the HEV oligo DNA standard curve was leveled off at high Ct values due to low input DNA concentrations. Bowers and Dhar also reported it; however, this effect was apparent when the plasmid DNA and 500 bases in vitro transcribed RNA template were used as templates. The data from their study suggested that the size of the template used to generate standard curve should be very similar to the size of the amplicon which will reduce or eliminate the nonspecific amplification. In this report, we use this strategy but we could not observe similar results. Nevertheless, later data demonstrated that this effect did not have a directly influence on HEV genome absolute quantification due to the exclusion of non-specific amplification templates.
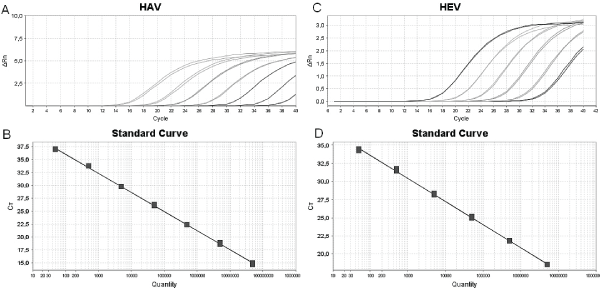
Figure 1: Synthetic oligo DNA standard curves for absolute quantification of hepatitis A and E virus by real-time PCR. Amplification plot and linear regression of HAV synthetic standard curve, respectively, slope -3,499, R2 0,99 (A) and (B). Amplification plot and linear regression of HEV synthetic standard curve, respectively, slope -3,301, R20,999 (A) and (B).
View Figure 1
Following, it was assayed both plasmid and synthetic DNA curves with the panel samples. As the plasmid DNA curve was optimized for both viral genomes (range 2x10-2-10-8ng/ul), they were used as "gold standard" curve to compare sample quantification for HAV and HEV. Samples quantification using each type of curve as standard was compared each other and the difference between their results were analyzed. HAV results demonstrated that quantification ratio between the two types of curves ranged from 0.83 to 2.14 times. The same comparison was performed for HEV samples and quantification ratio ranged from 1.87 to 2.23 times. Difference in absolute quantification between both types of curves, for HAV and HEV genomes, did not exceed one log. Furthermore, there was not discordance between qualitative results of tested samples.
Amplification efficiency values were similar for both types of curves, as indicated by the slopes of separate standard curves. The results of HAV absolute quantification demonstrated a slope average for plasmid DNA of -3,345, and oligo DNA of -3,499. For HEV, the slope average for plasmid DNA was -3,27 and for oligo DNA -3,301 (R20,99 for all assays).
The absolute quantification of viral genomes by real time PCR has been become routine. Conventionally, a standard curve is generated using a dilution series of cloned genomic segment of the target virus as template, which is time consuming, and costly [6].
Recently, oligonucleotide representing the target gene has been used as template to generate a standard curve, as reported in this study and ISO/TS 15216-1:2013 that describe a method for quantification of levels of HAV from test samples of foodstuffs or food surfaces.
Our RT-PCR quantification methods were not compared with ISO/TS 15216-1:2013. However, the real-time PCR to hepatitis A, described here, have been used since 2007 to detect and quantify HAV from water, food, saliva, cell culture, serum, liver and stool samples with plasmid standard curve [10-15]. The results showed that the standard curve can be replaced for a synthetic curve.
The synthetics biology can overcome the biological limitations of plasmid curve. We could demonstrate that oligo DNA could be used as template of standard curve due to its similar results with the plasmid DNA curves (gold standard). Oligo DNAs standard curves could be a helpful alternative or laboratories that do not have enough space and financial conditions for molecular cloning. Due to these characteristics, they could be a very useful tool on viral diagnosis.
Financial Support
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). PAPES VI.
References
-
Vaughan G, Goncalves Rossi LM, Forbi JC, de Paula VS, Purdy MA, et al. (2014) Hepatitis A virus: host interactions, molecular epidemiology and evolution. Infect Genet Evol 21: 227-243.
-
Arends JE, Ghisetti V, Irving W, Dalton HR, Izopet J, et al. (2014) Hepatitis E: An emerging infection in high income countries. J Clin Virol 59: 81-88.
-
Metcalf TG, Melnick JL, Estes MK (1995) Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology-a trip of over 50 years. Annu Rev Microbiol 49: 461-487.
-
Lee HK, Jeong YS (2004) Comparison of total culturable virus assay and multiplex integrated cell culture-PCR for reliability of waterborne virus detection. Appl Environ Microbiol 70: 3632-3636.
-
ISO/TS 15216-1: (2013) Microbiology of food and animal feed - Horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR - Part 1: Method for quantification.
-
Bowers RM, Dhar AK (2011) Effect of template on generating a standard curve for absolute quantification of an RNA virus by real-time reverse transcriptase-polymerase chain reaction. Mol Cell Probes 25:60-64.
-
Arkin AP, Schaffer DV (2011) Network news: innovations in 21st century systems biology. Cell 144: 844-849.
-
De Paula VS, Diniz-Mendes L, Villar LM, Luz SL, Silva LA, et al. (2007) Hepatitis A virus in environmental water samples from the Amazon Basin. Water Res 41: 1169-1176.
-
Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR (2006) A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods 131: 65-71.
-
de Paula VS, Perse AS, Amado LA, de Morais LM, de Lima SM, et al. (2009) Kinetics of hepatitis A virus replication in vivo and in vitro using negative-strand quantitative PCR. Eur J Clin Microbiol Infect Dis 28: 1167-1176.
-
de Paula VS, Gaspar AMC, Villar LM (2010) Optimization of Methods for Detecting Hepatitis A Virus in Food. Food and Environmental Virology. 2: 47-52.
-
Villar LM, de Paula VS, Diniz-Mendes L, Guimarães FR, Ferreira FF, et al. (2007) Molecular detection of hepatitis A virus in urban sewage in Rio de Janeiro, Brazil. Lett Appl Microbiol 45: 168-173.
-
Lima LR, De Almeida AJ, Tourinho Rdos S, Hasselmann B, Ximenez LL, et al. (2014) Evidence of hepatitis A virus person-to-person transmission in household outbreaks. PLoS One 9: e102925.
-
Amado LA, Marchevsky RS, de Paula VS, Hooper C, Freire Mda S, et al. (2010) Experimental hepatitis A virus (HAV) infection in cynomolgus monkeys (Macaca fascicularis): evidence of active extrahepatic site of HAV replication. Int J Exp Pathol. 91:87-97.
-
Amado LA, Villar LM, de Paula VS, Pinto MA, Gaspar AM (2011) Exposure to multiple subgenotypes of hepatitis A virus during an outbreak using matched serum and saliva specimens. J Med Virol 83: 768-775.