Early diagnosis and treatment of tuberculosis (TB) patients are the most effective ways to protect society from this disease. The objectives of this study were to evaluate the molecular epidemiology of mycobacterial strains by repetitive sequence PCR (rep-PCR) using the DiversiLab System method at Kayseri in Turkey.
A total of fifteen clinically sample isolated Mycobacterial strains were identified as M. tuberculosis complex (MTBC) and non-tuberculous mycobacteria (NTM) by the rep-PCR method (bioMerieux), SD (Standard Diagnostics) rapid tests. Genotyping was performed by the rep-PCR method and clonal affinity was researched.
Nine of the strains were identified as MTBC and six strains were identified as NTM by the SD rapid test methods. Five of the strains were identified as NTM by the rep-PCR method as two M. scrofulaceum, M. chelonae, M. fortuitum and M. avium complex (MAC). By genotyping with rep-PCR a total of nine clones were identified, which included two main clones (A and B) and seven other clones (C, D, E, F, G, H, I).
It was detected that rep-PCR test is useful method and can be used for the molecular epidemiology and for the identification of mycobacterial strains as they are rapid and practical. However, it is necessary to perform new studies for more mycobacterial strains using the methods described herein.
M. tuberculosis complex, Non-tuberculous mycobacteria, Genotyping, Repetitive sequence PCR by DiversiLab System, SD bioline TB Ag MPT64 rapid identification test
Tuberculosis (TB) is a life-threatening infectious disease which continues to maintain its significance worldwide with the emergence of resistant strains. There were new cases with resistance to rifampicin (RRTB) and multidrug-resistant TB (MDR-TB) in cases of TB recently [1].
Early detection of the disease caused by members of the M. tuberculosis complex (MTBC) is essential for initiating the appropriate therapy and controlling the transmission of tuberculosis. Apart from the members of the MTBC and non-tuberculous mycobacteria (NTM), species are increasing in number and some of them are sources of important diseases in humans. Thus, in order to decrease the tuberculosis mortality rate has rapid and reliable identification of MTBC and NTM is essential. This should be done by including the use of more rapid and efficient laboratory algorithms to detect M. tuberculosis and NTM and susceptibility testing against antituberculosis drugs [1,2].
Conventional phenotypic identification methods are the current "gold standard" for assessing the identification of MTBC and NTM. Rapid, accurate diagnosis of MTBC and NTM is critical for timely initiation of treatment, and control of the diseases. Apart from the members of the MTBC and NTM, species are increasing in number and some of them are sources of important diseases in humans [2-4]. Another differentiating factor between MTBC and NTM is the distinctive chording that molecular methods including RealTime MDR TB (Abbott Molecular) [5], and BDProbeTec (Becton Dickinson). Alternatively, molecular assays provide a faster turnover time while maintaining high sensitivity and specificity. Quantitative real-time polymerase chain reaction (rep-PCR) is considered as one of the most widely applied molecular assays for the diagnosis of tuberculosis [6].
Epidemiological studies are important for mycobacterial outbreaks and for protection against them. Therefore, epidemiological studies should be given importance [7-9].
The present study was conducted to assess rep-PCR for rapid differentiation between MTBC and NTM and the molecular epidemiology of Mycobacterial strains in clinical isolates at Kayseri in Turkey.
A total of 15 Mycobacterial strains, which were isolated from 15 patients at the time of diagnosis, were studied in the Clinical Microbiology Department of Gevher Nesibe Hospital of Erciyes University in Kayseri, Turkey (Central Anatolia) over a period of one year. These samples were from bronchoalveolar lavage (n = 3), sputum (n = 5), urine (n = 3), pleura (n = 3), and gastric lavage fluid (n = 1).
Positive vials obtained by the MGIT 960 TB culture system and colony on Lowenstein-Jensen medium were subjected to smear microscopy for the presence of acid-fast bacilli (AFB). The SD Bioline MPT64 Ag test (SD), an immunochromatographic assay using mouse monoclonal antibodies to detect MPT64 antigen/protein, which is specific for M. tuberculosis complex, was used in combination with culture methods to distinguish TB and NTM as a routine reference method. The performances of rep-PCR were evaluated to compare with SD, by using a combination of the MGIT 960 TB culture system and SD tests as the "gold standard" [10,11].
The genetic proximity of 15 Mycobacterium strains was determined by using the rep-PCR method.
For all isolates cultured on SDA, DNA was extracted using the UltraClean Microbial DNA isolation kit (Mo Bio Laboratories, Solana Beach, Calif.) following the manufacturer's instructions. The presence of high-molecular-mass DNA was confirmed by agarose gel electrophoresis.
For all isolates, DNA was amplified using a Diversilab Mycobacterium Kit (BioMerieux, France) for DNA fingerprinting, in accordance with the manufacturer's instructions. The thermal cycling parameters were as follows: initial denaturation at 94 ℃ for 2 min, 35 cycles of denaturation at 94 ℃ for 30 s, annealing at 50 ℃ for 30 s, extension at 70 ℃ for 90 s, and a final extension at 70 ℃ for 3 min.
Analysis of PCR products was implemented using a microfluidics chip with an Agilent 2100 Bioanalyzer (AgilentTechnologies, Calif., USA). For all isolates, the rep-PCR-based fingerprint patterns were obtained in gel-like images by using DiversiLab 2.1.66 web-based interpretation software. For analysis, the Pearson correlation coefficient was used in the similarity calculation and UPGMA was used to automatically compare the rep-PCR profiles.
Species identification of the fungal cultures was based on their cluster similarity percentages obtained from the dendrograms, as well as a visual comparison of the virtual gel images. In this study, a culture was given species identification if the fingerprint pattern had ≥ 85% similarity to a single fungus or a cluster of fungi in the database. Isolates with a similarity coefficient equal to or greater than 95% were evaluated as main clones. Those with a similarity coefficient equal to or greater than 95% and less than 97% were classified as sub-clones, and isolates showing a similarity coefficient of less than 95% were identified as different clones [12,13].
In our study, mycobacterial strains isolated from positive cultures were identified by two different methods namely the SD rapid test and rep-PCR methods for the 15 isolated mycobacterial strains, SD rapid test methods were used and 9 strains were identified as MTBC and 6 strains as NTM. For rep-PCR methods, using SD as the gold standard the sensitivity, specificity, were 93.3% and 100% respectively (Table 1).
Table 1: Diagnostic evaluation of SD testing and Rep-PCR. View Table 1
In the identification at species level using the rep-PCR method in 15 strains that were positive for mycobacteria, 10 were identified as M. tuberculosis and 5 as NTM. Three of the NTM strains were identified as M. chelonae, M. fortuitum, Mycobacterium avium complex (MAC) and the other two were identified as M. scrofulaceum.
A total of 9 clones were identified by rep-PCR analysis, with two main clones (A, B) and seven different clones (C, D, E, F, G, H, I). A gel-like image, percentage similarity ratio, and dendrogram of the Mycobacterium isolates were formed by the rep-PCR method with DiversiLab software, version 3.3. and can be seen in Figure 1.
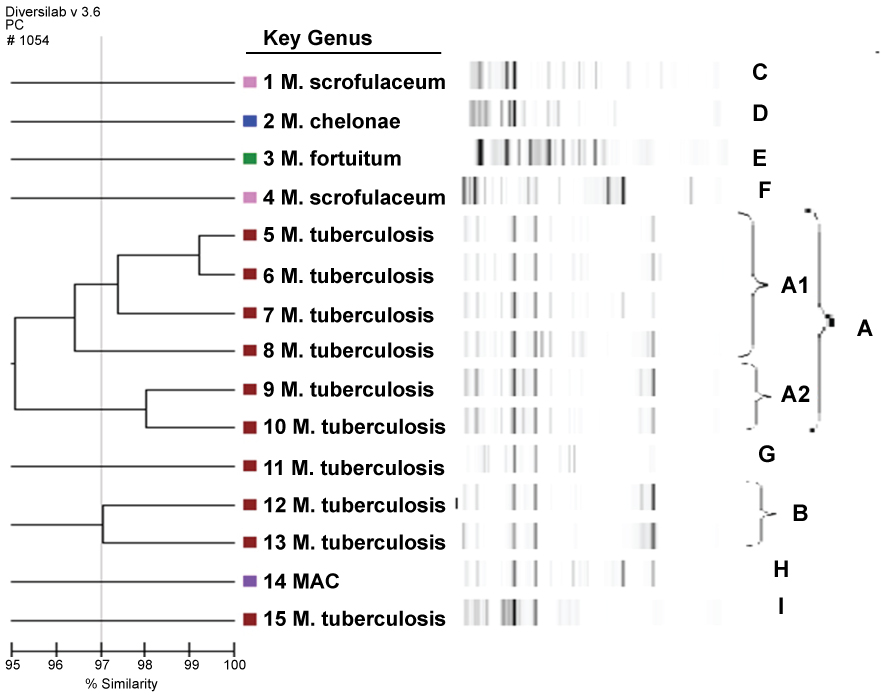 Figure 1: Rep-PCR-based sim. View Figure 1
Figure 1: Rep-PCR-based sim. View Figure 1
The dendrogram and gel-like image graph of 15 isolated mycobacterial strains are shown in Figure 1. Clone A was identified as the predominant type among the mycobacterial strains and 6 strains (33.3%) were found to belong to clone A. All of these strains were identified as MTBC with the BD rapid test method. Two different subtypes (A1-A2) of clone A were identified. It was seen that 4 strains belonging to the A clone belonged to were identified from the A1 clone and 2 strains belonged to the A2 clone. The strains belonging to the A1 clone two sputum, a pleura and a urine sample sent from three chest disease patients and one from the urogenital polyclinic. One of the A2 clones was identified from two urine samples sent from the Urology and Infectious Diseases Polyclinic. The dendrogram and gel-like image of clone A are shown in Figure 1.
Clone B was identified as the second major clone in mycobacterial strains. Both strains identified as belonging to clone B were identified with the SD rapid test and BD rapid test methods as MTBC. MTBC is composed of strains isolated from the sputum specimens in the same period sent to the Mycobacteriology laboratory from the Department of Clinical and Chest Diseases of Hematology. The dendrogram and gel-like image of clone B are shown in Figure 1. In mycobacterial strains, 7 clones of 7 different strains with similarity rates of less than 95% were identified except for 2 major (A-B) clones.
Clinical presentation of Mycobacterium tuberculosis complex (MTBC) and non-tuberculous mycobacteria (NTM) infections may be similar, but treatment is always different. NTM are inherently resistant to conventional anti-tuberculosis drugs, hence often misdiagnosed as multidrug-resistant tuberculosis. With the increasing number of clinical NTM infections, accurate differentiation between MTBC and NTM is crucial [2].
NTM is regarded as an increasingly important cause in lung disease. In particular, some NTM species, especially MAC members, are frequently isolated from clinical laboratories. Molecular methods have also found wide application in the identification of mycobacteria in clinical specimens and at species level [14,15]. Koh, et al. [16] in 2006, isolated NTMs from respiratory tract specimens which were defined mainly as MAC (48%) and M. abscessus (33%). In a study conducted by the Korean National Tuberculosis Association, NTMs were identified and were found as follows 65.2% M. avium-intracellulare, 12.7% M. fortuitum, 9.5% M. chelonae, 4.4% M. gordonae, 3.2% M. terrae, 1.8% M. scrofuloceum, 1.3% M. kansasii and M. szulgai [11]. In the study by Couto, et al. [14], NTMs isolated from clinical samples [15] were identified mainly as follows 24.8% MAC, 15.4% M. gordonae, 9.4% M. fortuitum, 6.7% M chelonae, 6% M. xenopi, 4.7% M. peregrinum and 4% M. abscessus. In our study, 66.7% of cases were found to be M. tuberculosis by the rep-PCR method. Two strains identified as NTM were identified as M. scrofulaceum while the others were identified as MAC, M. fortuitum, and M. chelonae. We have evaluated only 15 strains, and this is a limitation of my study.
When the epidemiological data point to a TB epidemic, genotyping methods reveal whether it is a real epidemic, or just a lot of cases that have coincidentally come together. According to these results, the spread of infection can be prevented by evaluating the community health parameters. Genotyping methods are used to determine disease source and transmission time. Genotyping, which provides information about infection, is needed for epidemiological and clinical purposes with TB control [8,15]. Genotypic analysis of M. tuberculosis isolates has been implemented to support TB epidemic control activities. It has been further facilitated by PCR-based typing methods that provide genetic characterization of samples containing a small number of TB bacilli [9]. The M. tuberculosis DNA fingerprint is a powerful tool for molecular epidemiological investigations. Standard M. tuberculosis fingerprinting methods include spoligotyping, IS6110 RFLP analysis, MIRU and rep-PCR methods [17,18].
IS6110 RFLP is accepted to be the most commonly used genotyping method for M. tuberculosis molecular epidemiology. Repeated sequence-based PCR (rep-PCR) as an epidemiological tool has been recognized as an effective method. Rep-PCR is used to determine the resemblance of bacterial strains at genomic level, such as repeating sequences throughout the genome that can discriminate between mutations based on amplicon density and size. Rep-PCR has developed strain discrimination and test repeatability when compared to PCR-based platforms. The commercial DiversiLab Mycobacterial Typing System (Biomeriux) has the advantages of real-time data production and partial automation ease [8]. Masala, et al. [12] compared the M. tuberculosis isolate IS6110 RFLP, Mycobacterial Interspersed Repetitive Unit Variable Number Tandem Repeat (MIRU-VNTR) typing and rep-PCR methods in a study conducted in Italy. Four major clones of the strains were identified by MIRU-VNTR typing and the same isolates were joined by RFLP typing. With the rep-PCR method, a very different fourth clone was identified with only three of these major clusters. At the end of their study, the authors reported that the DiversiLab rep-PCR system is very expensive, and requires trained staff in spite of its integrated, automatic and rapid testing platform. In a similar study using NTM isolate, the authors compared the rep-PCR with RAPD (Random Amplification of Polymorphic DNA), hsp65 gene sequencing and MIRU-VNTR methods and found that the rep-PCR method is suitable for any Mycobacterium species and can be completed with a small amount of DNA work but has been reported to provide lower levels of discrimination for conditions such as epidemiological surveys [19]. In another study conducted on immigrants in Switzerland, isolates of M. tuberculosis obtained from 15 TB cases originating from the same country were analyzed by RFLP and rep-PCR. RFLP analysis 10 and rep-PCR method 11 detected the M. tuberculosis isolate with the same genotype and has revealed an active outbreak in a specific group of immigrants in Geneva [19]. In a study conducted after a major TB epidemic in King County, Washington, in the USA in homeless people, genotyping was performed by the rep-PCR method to rapidly detect new cases including a strain called SBR19. While SBR19, an outbreak strain, was not seen before 2002 in this region, it was identified in 53% of cases in 2003, 77% in 2003, 83% in 2004 and 40% in the first half of 2005. This epidemic strain continued to manifest itself throughout the study within the homeless population. It was reported in this study that rep-PCR is very fast and cost effective and facilitates public health work by preventing new outbreaks [18]. Jang, et al. [8] conducted a study of 89 the M. tuberculosis isolates between 2008 and 2009 and assessed the ability of IS6110 RFLP data and the DiversiLab rep-PCR system to differentiate. Using the IS6110-RFLP method, 76 isolates were defined as the Beijing strain, and 13 isolates were defined as non-Beijing. When a 95% similarity level was applied, these 89 isolates were identified by the DiversiLAB system as a total of 8 clones together with the main clone consisting of 18 isolates and a separate isolate. The authors reported that the DiversiLAB system could be used in epidemiological studies of M. tuberculosis and was capable of a high degree of distinction, and produced repeatable, standardized and precise results. In our study, a total of 9 clones consisting of two main clones (A, B) and seven different clones (C, D, E, F, G, H, I) were identified by rep-PCR analysis. Six strains (33.3%) belonged to clone A and this was the predominant type. All of these strains were typed as M. tuberculosis by the rep-PCR method and as MTBC by the SD rapid test methods. Two distinct subtypes (A1-A2) were identified in clone A and 4 strains belonging to clone A were found to belong to A1 clone and 2 strains to belong to A2 clone. In our study, clone B was identified as the second clone, and both strains belonging to this clone were typed as M. tuberculosis by the rep-PCR method and as MTBC by the SD rapid test methods. This clone consisted of strains isolated from sputum and BAL samples sent to the Mycobacteriology Laboratory in recent history.
As a result, identification of mycobacteria as species level is important in terms of clinical approach and in the regulation of treatment in TB patients. The rep-PCR method was able to identify mycobacterial strains easily and quickly at species level. For this reason, the rep-PCR method appears to have the advantage in mycobacteria when evaluating both the species identification level and the clonal approach due to the ease of automation, the accessible database and the ease of repetition. However, new studies should be carried out with a higher number of strains sand research needs to continue on these subjects.
We would like to also thank Biologist Gonca Demir, Dr. Hafize SAV and Dr. Humen Cebbari (BMLabosis) for helping with the laboratory studies.
This work with the number: TSA-11-3642; was supported by the Department of Scientific Research Projects (BAP) Commission, Erciyes University, Kayseri, Turkey.
We declare that we have no conflict of interest. The authors alone are responsible for the content and the writing of the article.
This work with the number:TSA-11-3642; was supported by the Department of Scientific Research Projects (BAP) Commission, Erciyes University, Kayseri, Turkey.
Murat KARAUZ's contribution in this manuscript; in the collection and laboratuar studies of Mycobacterium isolates in this study. He is "Laboratuar investigator" .
A. Nedret KOÇ's contribution in in study design; analysis and interpretation of data; in the writing of this manuscript; and in the decision to submit the article for publication. She is "critically reviewed the study proposal'' in this study.
M. Altay ATALAY's contribution in this manuscript; in study design; in analysis and interpretation of data. He is "participating investigator".