International Journal of Neurology and Neurotherapy
Association between the ERCC1 Polymorphisms and Glioma Risk: A Meta-Analysis of Case-Control Studies
LIU Yan1#, CAI Xiao Qin2#, ZHAO Lian Ying3, SHEN Heng Shan2, HU Jian Wei4,*
1Department of Epidemiology, School of Public Health, Medical College of Soochow University, Suzhou, Jiangsu, China
2Department of Diagnostic Center, the Kunshan Affiliated Hospital of Nanjing University of Traditional Chinese Medicine, Suzhou 215300, Jiangsu, China
3Health Supervision Institute of Kunshan, Kunshan, Jiangsu, China
4Maternal and Child Health Bureau of Kunshan, Kunshan, Jiangsu, China
#These authors contributed equally to this work and should be considered as co-first authors.
*Corresponding author:
HU Jian Wei, Physician, Maternal and Child Health Bureau of Kunshan, Kunshan, Jiangsu, China, Tel: 086-512-5733-6557; E-mail: hujianwei19850826@163.com
Int J Neurol Neurother, IJNN-2-034, (Volume 2, Issue 2), Original Research; ISSN: 2378-3001
Received: June 01, 2015 | Accepted: October 10, 2015 | Published: October 13, 2015
Citation: LIU Yan, CAI Xiao Qin, ZHAO Lian Ying, SHEN Heng Shan, Wei HUJ (2015) Association Between the ERCC1 Polymorphisms and Glioma Risk: A Meta-Analysis of Case-Control Studies. Int J Neurol Neurother 2:034. 10.23937/2378-3001/2/2/1034
Copyright: © 2015 LIU Yan, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Objective: Polymorphisms in DNA repair genes have been shown to influence DNA repair processes and to modify cancer susceptibility. Published data regarding the association between excision repair cross-complementing rodent repair deficiency complementation group1 (ERCC1) polymorphisms and glioma risk have been inconsistent and inconclusive. To acquire a more precise effect of the association between these polymorphisms and glioma risk, a meta-analysis was performed.
Methods: Data was collected in PubMed and EMBASE, with the last search up to 30th August 2013. A total of 6 studies were identified with 2642 cases and 3669 controls for ERCC1 C8092A polymorphism, and 4 studies were identified with 1390 cases and 1546 controls for ERCC1 C118T polymorphism. All of the statistical analyses were performed using statistical data 10.0.
Results: The combined results showed that ERCC1 C8092A polymorphism was associated with glioma risk (additive model: OR = 1.10, 95%CI 1.02 - 1.20; recessive model: OR = 1.51, 95 % CI 1.24 - 1.85; co-dominant model AA vs. CC: OR = 1.52, 95 % CI 1.24 - 1.86). As for ethnicity subgroup analysis, ERCC1 C8092A polymorphism was associated with increased glioma risk among Chinese(additive model: OR = 1.15, 95 % CI 1.01 - 1.30; recessive model: OR = 1.34, 95 % CI 1.02 - 1.75; AA vs. CC: OR = 1.37, 95 % CI 1.03 - 1.81), and so among Caucasian except additive model(recessive model: OR = 1.75, 95 % CI 1.31 - 2.34; co-dominant model AA vs. CC: OR = 1.70, 95 % CI 1.27 - 2.29). No evidence of an association of ERCC1 C118T polymorphism with glioma was found.
Conclusion: The meta-analysis suggested that ERCC1 C8092A polymorphism might be associated with the increased risk of glioma, whereas ERCC1 C118T polymorphism might have no influence on the susceptibility of glioma.
Keywords
Polymorphism, ERCC1, Glioma, Meta-analysis
Introduction
Glioma is the most common and fatal neurological cancer [1,2] and accounts for almost 80% of primary malignant brain tumors [3,4]. Despite many advances in surgical and medical therapy in recent years, glioma consistently remains a fatal disease and there is no significant increase in the survival for patients with glioma [1,2]. Though several risk factors have been found, the exact pathogenesis of glioma remains unclear [5]. However, there is no doubt that genetic factors play important roles in the development of glioma [6,7]. Numerous studies are addressing associations of polymorphisms in DNA repair genes and cancer risk [8] because accurate and efficient DNA repair is crucial to genomic integrity and fidelity. The DNA repair system is complex, governed by more than 125 genes, many of which are polymorphic [9-12]. The DNA repair gene, excision repair cross-complementing rodent repair deficiency complementation group1 (ERCC1), whose products are important in the process of nucleotide excision repair (NER) lies on chromosome 19q13.3 in a putative glioma suppression region [13]. ERCC1 gene is reported to be a crucial gene in the NER pathway, and ERCC1 polymorphisms can modify the function of NER pathway, thus influence the risk of human cancers [14]. ERCC1 C8092A and C118T are two common polymorphisms of ERCC1 gene and they alters risk of several types of cancers [15-18]. Associations between polymorphisms in ERCC1 gene and glioma risk have also been examined but the observed associations were inconsistent [19-22], and a single study may be too underpowered to detect a possible small effect of the polymorphisms on glioma, especially when the sample size was relatively small. To the best of our knowledge, there has been no comprehensive quantitative assessment of the association of ERCC1 C8092A or C118T polymorphism with glioma risk. Hence, we performed a meta-analysis of all eligible studies to derive a more precise estimation of the associations of ERCC1 C8092A or C118T polymorphism with glioma risk.
Materials and Methods
Literature search
PubMed and EMBASE were searched (the last search update on the 30th August 2013) using the search terms: 'ERCC1' and 'glioma or brain tumor'. All studies matching the eligible criteria were retrieved, and bibliographies checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were hand searched to identify additional studies. Only published studies with full text were included in this meta-analysis.
Inclusion criteria
The inclusion criteria were (a) evaluation of the ERCC1 C8092A or C118T polymorphism and glioma risk, (b) case-control study design, (c) human subject studies and (d) the size of the sample, odds ratios (ORs) and their 95% confidence intervals (CIs) or the information that can help infer the results in the papers.
Data extraction
Information was carefully extracted from all eligible publications independently by two investigators (Jun Liu and Ting Lai), according to the inclusion criteria listed above. The following data was collected from each study: first author's surname, publication date, country, ethnicity, study design, genotyping methods, total number of cases and controls, and numbers of cases and controls with ERCC1 C8092A and C118T genotypes, respectively. For those studies that included subjects of different ethnic groups, data were extracted separately for each of the ethnic groups, categorized as Caucasians or Chinese. We did not define any minimum number of patients for including a study in our meta-analysis. When the same patient population was included in several studies only the most recent or complete study was included in this meta-analysis.
Statistical analysis
Crude ORs with 95% CIs were calculated, according to the method of Woolf [23] to assess the association of ERCC1 C8092A and C118T polymorphisms with glioma risk. The pooled ORs for ERCC1 C8092A were performed for additive model (A allele vs. C allele), co-dominant model (AA vs. CC, AC vs. CC), dominant model (AA/AC vs. CC), recessive model (AA vs. AC/CC). As for ERCC1 C118T all these estimates were also calculated. Heterogeneity assumption was checked by a chi-square-based Q-test [24]. A p-value of more than 0.10 for the Q-test indicated a lack of heterogeneity across the studies, so the pooled estimation of the ORs of each study was calculated by the fixed effects model (Mantel-Haenszel method). Otherwise, the random effects model (DerSimonian and Laird method) was used [25]. The significance of the pooled OR was determined by the Z-test, and P < 0.05 was considered as statistically significant. To evaluate the ethnic-specific effect, subgroup analysis was conducted on the basis of different ethnicities. One-way sensitivity analysis was performed to assess the stability of the results, namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled OR [26,27]. An estimate of the potential publication bias was carried out by funnel plot, in which the standard error (SE) of log (OR) of each study was plotted against its log (OR). An asymmetric plot suggested a possible publication bias. The funnel plot asymmetry was assessed by Egger's test, a linear regression approach to measure funnel plot asymmetry on the natural logarithmic scale of the OR. The significance of the intercept was determined by the t-test suggested by Egger, P < 0.05 was considered representative of statistically significant publication bias [28]. Hardy-Weinberg equilibrium (HWE) in the control group was tested by the chi-square test for goodness of fit, and a p-value of < 0.05 was considered significant. All the statistical tests for our meta-analysis were performed with STATA version 10.0 (Stata Corporation, College Station, TX). All p-values were two-sided.
Results
Study characteristics
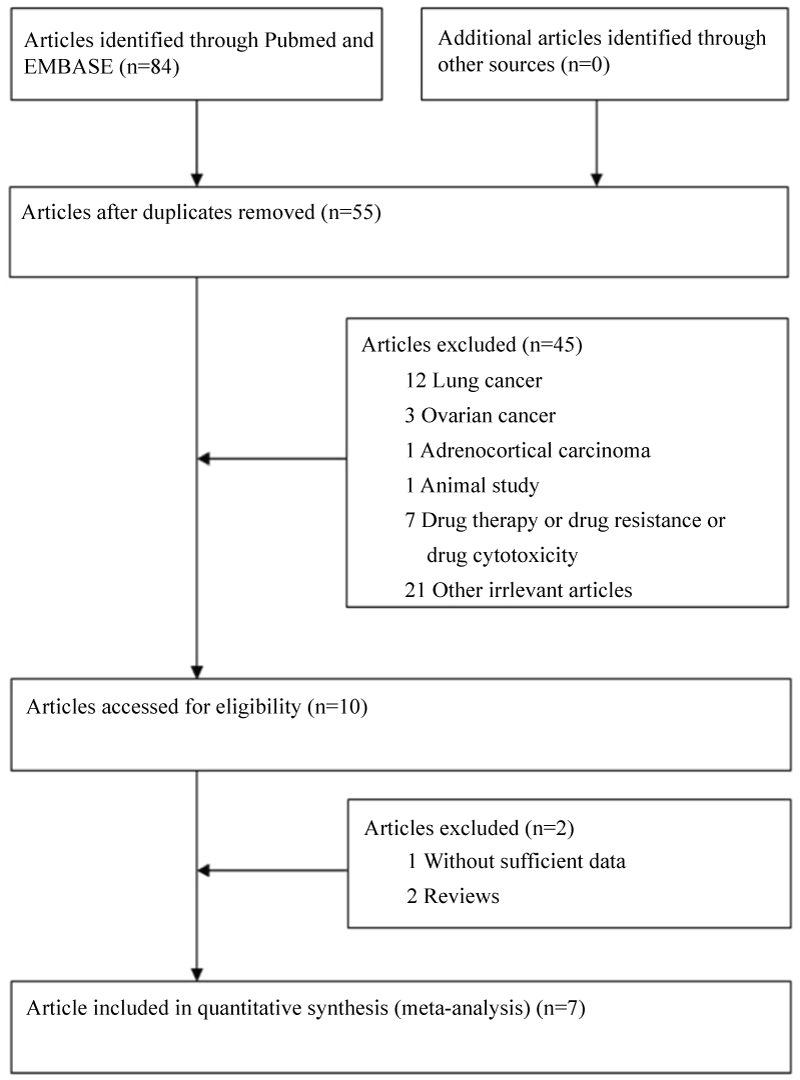
Based on the search criterion, 84 articles were found. Through the step of screening the articles (Figure 1), seven articles [20-22,29-32] were included. Characteristics of included studies are summarized in table 1. Among of these studies, a total of 6 studies involving 2642 cases and 3669 controls for ERCC1 C8092A, while 4 studies involving of 1390 cases and 1546 controls for ERCC1 C118T were analyzed. For ERCC1 C8092A, 3 studies were carried on Chinese population, and other 3 studies were carried on Caucasian population. For ERCC1 C118T, 4 studies were provided data on Chinese population. MassARRAY was used to validate genotype in two papers, and TaqMan was used to validate genotype in two studies. Mixed PCR method was used to validate genotype in two studies and PCR-SSCP was used in one study. Genotype distributions in the control populations were all in agreement with HWE.
![]()
Table 1: Main characteristics of all studies included in the meta-analysis.
View Table 1
Meta-analysis results
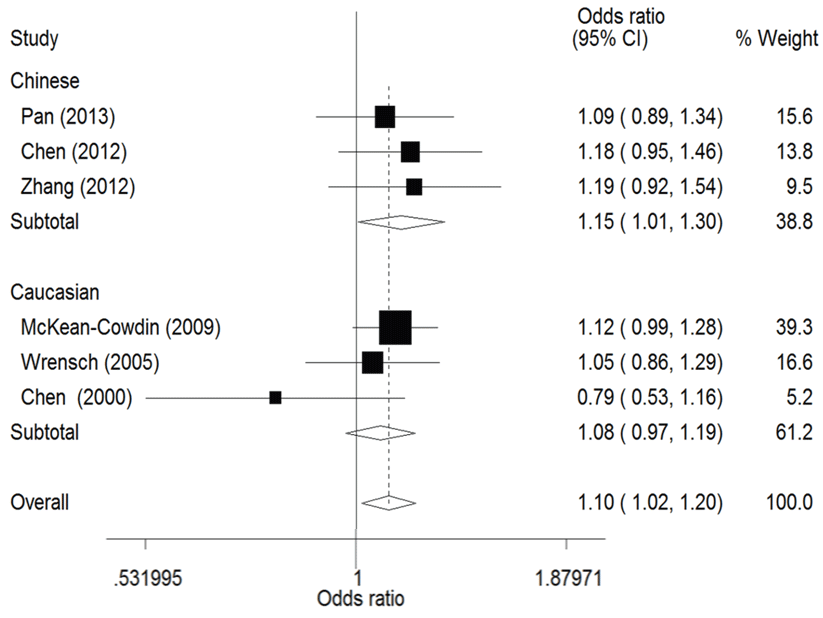
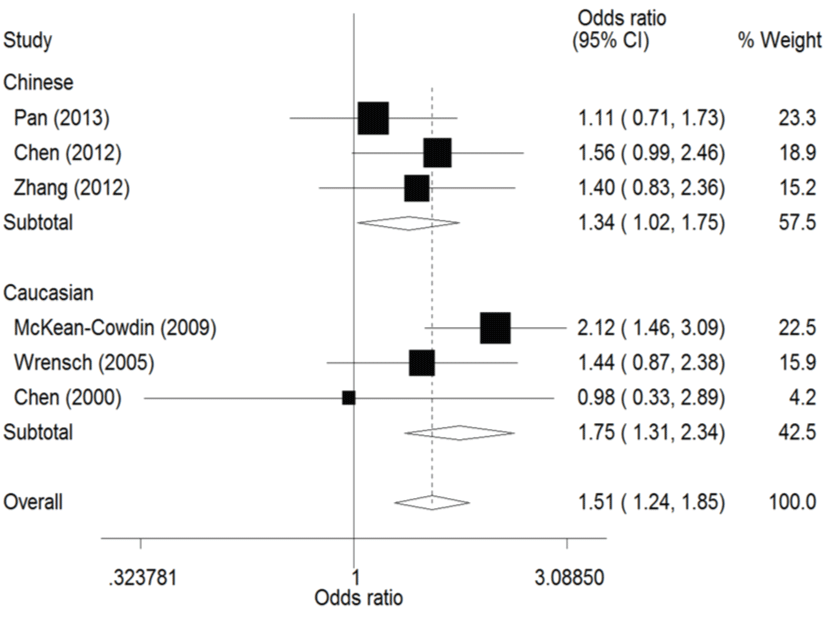
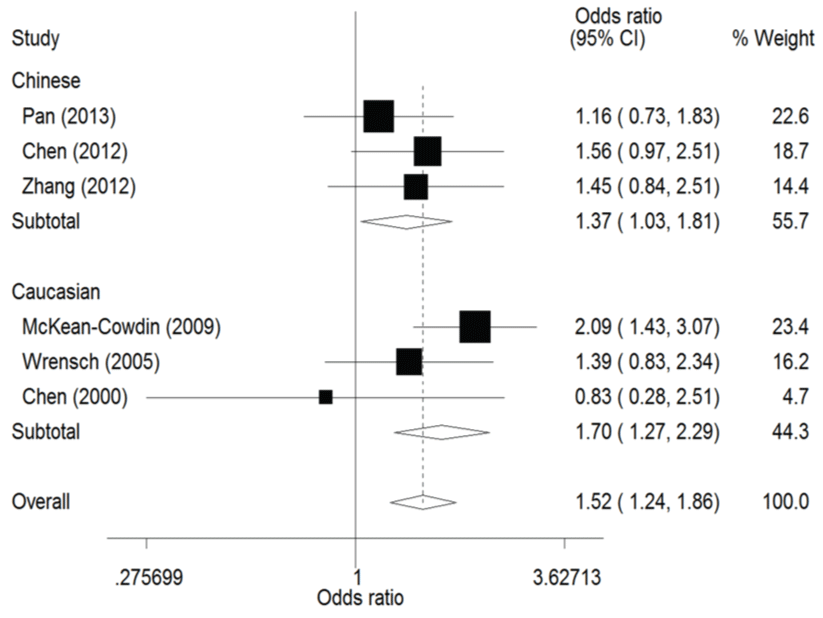
The result of this meta-analysis is shown in table 2. For ERCC1 C8092A polymorphism, Q-test was used in all of the genetic models and there was no significant heterogeneity. Therefore, the pooled ORs were calculated using fixed-effects model. No association for ERCC1 C8092A polymorphism and glioma risk was found in these genetic models (AA/AC vs. CC: OR = 1.04, 95% CI 0.94 - 1.06; AC vs. CC: OR = 0.98, 95% CI 0.88 - 1.09). Whereas there was significant association between ERCC1 C8092A polymorphism and susceptibility to glioma, which could be identified in these genetic models (A vs. C: OR = 1.10, 95% CI 1.02 - 1.20, figure 2; AA vs. AC/CC: OR = 1.51, 95% CI 1.24-1.85, figure 3, AA vs. CC: OR = 1.52, 95% CI 1.24-1.86, figure 4).
.
Figure 2: Forest plots of the odds ratios (ORs) and 95% confidence intervals (CIs) for ERCC1 C8092A polymorphism and risk of glioma observed in subgroup analysis by ethnicity (fixed effects model). (Additive model: A vs. C).
View Figure 2
![]()
Table 2: Results of meta-analysis for ERCC1 C8092A and C118T and the risk of glioma.
View Table 2
In the ethnicity subgroup analysis, as for Chinese population there was significant association between ERCC1 C8092A polymorphism and susceptibility to glioma in these genetic models (A vs. C: OR = 1.15, 95% CI 1.01 - 1.30, figure 2; AA vs. AC/CC: OR = 1.34, 95% CI 1.02 - 1.75, figure 3; AA vs. CC: OR = 1.37, 95% CI 1.03 - 1.81, figure 4). And in Caucasian population these genetic models were significant (AA vs. AC/CC: OR = 1.75, 95% CI 1.31 - 2.34, figure 3; AA vs. CC: OR = 1.70, 95% CI 1.27 - 2.29, figure 4).
.
Figure 3: Forest plots of the odds ratios (ORs) and 95% confidence intervals (CIs) for ERCC1 C8092A polymorphism and risk of glioma observed in subgroup analysis by ethnicity (fixed effects model). (Recessive model: AA vs. AC/CC).
View Figure 3
.
Figure 4: Forest plots of the odds ratios (ORs) and 95% confidence intervals (CIs) for ERCC1 C8092A polymorphism and risk of glioma observed in subgroup analysis by ethnicity (fixed effects model). (Co-dominant model: AA vs. CC).
View Figure 4
For ERCC1 C118T polymorphism, no heterogeneity was found among all of genetic models either. There was no significant association between ERCC1 C118T polymorphism and glioma risk, which could be identified in any of the genetic models (T vs. C: OR = 1.02, 95% CI 0.91 - 1.13; TT/CT vs. CC: OR = 1.04, 95% CI 0.89 - 1.22; TT vs. CT/CC: OR = 0.99, 95% CI 0.83 - 1.19; TT vs. CC: OR = 1.04, 95% CI 0.84 - 1.29; CT vs. CC: OR = 1.03, 95% CI 0.87 - 1.22).
Sensitivity analysis
We deleted one single study from the overall pooled analysis each time to check the influence of the removed data set to the pooled ORs. No study was observed to change the homogeneity in heterozygote comparison.
Bias diagnostics
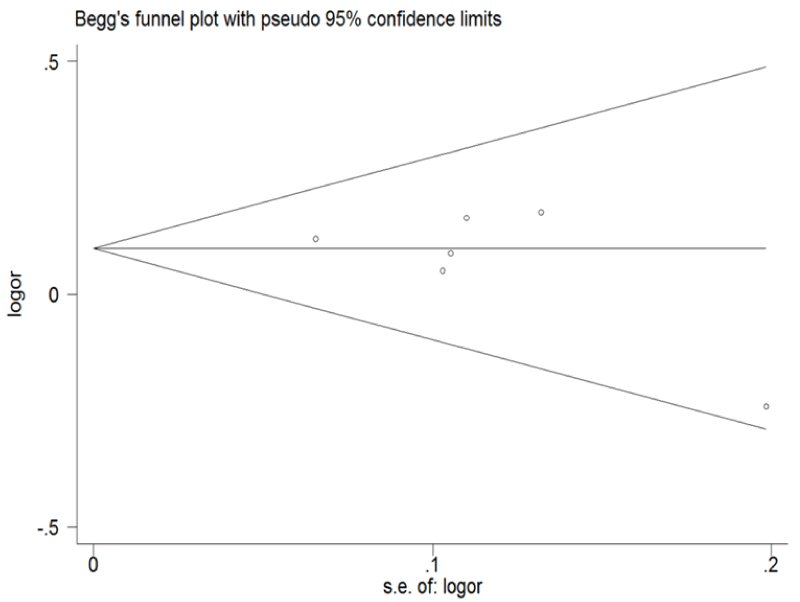
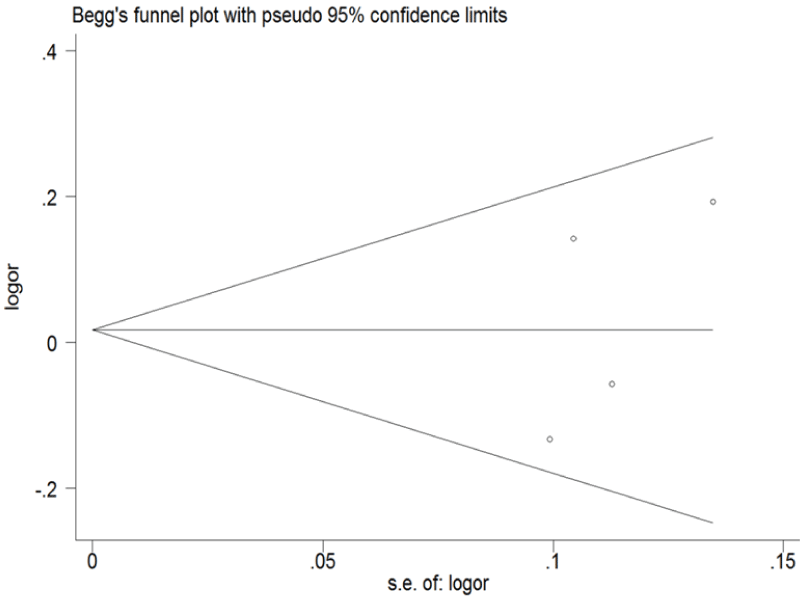
Funnel plot and Egger's test were performed to assess the publication bias of the literature (Figure 5 and Figure 6). Symmetrical funnel plots were obtained in ERCC1 C8092A and C118T tested in all of the models. Egger's test further confirmed the absence of publication bias in this meta-analysis (P > 0.05). No evidence of publication bias was observed in any comparison model.
.
Figure 5: Begg's funnel plot of publication bias for ERCC1 C8092A and C118T. (Begg's funnel plot for C8092A: A vs. C). Each point represents a separate study for the indicated association; horizontal line represents the meta-analysis summary estimate; log OR, natural logarithm of OR; s.e. of logOR, standard of the log OR.
View Figure 5
.
Figure 6: Begg's funnel plot of publication bias for ERCC1 C8092A and C118T. (Begg's funnel plot for C118T: T vs. C). Each point represents a separate study for the indicated association; horizontal line represents the meta-analysis summary estimate; log OR, natural logarithm of OR; s.e. of logOR, standard of the log OR.
View Figure 6
Discussion
The NER is a highly conserved DNA repair pathway that repairs DNA lesions which alter the helical structure of the DNA molecule and interfere with DNA replication and transcription [33]. The NER pathway repairs bulky lesions such as pyrimidine dimers, other photo-products, larger chemical adducts, and cross-links. The NER pathway involves at least four steps: (a) damage recognition by a complex of bound proteins including XPC, (b) unwinding of the DNA by the TFIIH complex that includes XPD, (c) removal of the damaged single-stranded fragment (usually about 27 - 30 bp) by molecules including an ERCC1 and XPF complex, and (d) synthesis by DNA polymerases [8]. The NER pathway removes damaged DNA bases by introducing nicks 5' and 3' to an abasic site in vitro, and NER contributes to the release of 8-oxoguanine from DNA [34,35]. Variation in efficiency of these processes might influence either cancer development.
In recent years, interest in the genetic susceptibility to cancers has led to a growing attention to the study of polymorphisms of genes involved in tumourigenesis. The DNA repair gene, excision repair cross-complementing rodent repair deficiency complementation group1 (ERCC1), is potentially relevant to cancer because of their involvement in the process of nucleotide excision repair (NER) [8]. The ERCC1 gene has been widely studied, and the ERCC1 C8091A TT genotype was observed to increase the risk of lung cancer [36]. There have been a few studies reported the relationship between the ERCC1 C8092A polymorphism and glioma susceptibility [19-22,30-32]. And only a few of these studies showed the ERCC1 C8092A polymorphism was associated with the risk of glioma [21], however, some not [19,20,22,30-32]. In the present meta-analysis, the combined results based on all studies showed that ERCC1 C8092A polymorphism was significantly associated risk (additive model: OR = 1.10, 95% CI 1.02 - 1.20; recessive model: OR = 1.51, 95% CI 1.24 - 1.85; AA vs. CC: OR = 1.52, 95% CI 1.24 - 1.86). The results indicated that individuals carrying at least one T allele might be at increased risk of glioma, which in agreement with the previous study, which suggested that the T allele may serve as a dangerous biomarker [21]. Using data collected through 1994, Chen found a statistically significant association with oligoastrocytomas (OR = 4.6; 95% CI, 1.6 - 13.2), but not with other types of glioma [21]. According to described above, our finding suggested that ERCC1 C8092A polymorphism may play a finite role on glioma risk. More studies and larger samples will be needed to further identify this relationship.
Although many studies have been performed to explore the etiology of glioma, the etiology is still not completely understood. Glioma, as a complex disease, is considered as a result of combined effects of multi-factors, including the inherited and environmental factors. Unfortunately, there are few studies focused on the gene-environment and gene-gene interactions with glioma risk. A significant and protective effect was found by Roberta when three single-nucleotide polymorphisms (ERCC2 rs13181, ERCC1 rs3212986, and GLTSCR1 rs1035938) located near each other on chromosome 19 were modeled as a haplotype. The most common haplotype (AGC) was associated with a 23% reduction in risk (P = 0.03) compared with all other haplotypes combined [22]. So, further studies should include the gene and environmental factors and detect the potential interactions between ERCC1 C8092A polymorphism and these factors.
In our study, we did not find any relation between the risk of glioma and ERCC1 C118T polymorphism, may either due to the minor effect between ERCC1 C118T polymorphism and glioma risk or to the relatively small sample size, and that is consistent with many previous studies [29-32].
To the best of our knowledge, this is the first meta-analysis evaluating the potential association between two common polymorphisms ERCC1 C8092A and C118T and the susceptibility to glioma. However, some limitations of this meta-analysis should be acknowledged. Firstly, our finding conflicts with some articles which indicated that there was no statistical evidence to assume a correlation between ERCC1 C8092A polymorphism and glioma risk. The reason is that one of the six studies [22] included in our meta-analysis related to ERCC1 C8092A played an important role in the pooled OR. According to the data provided in the study, the OR was calculated (recessive model: AC vs. AC/CC, OR = 2.09, 95% CI 1.43 - 3.07), that indicated the ERCC1 C8092A TT genotype increased glioma risk. However, after adjusted for age, gender, and study center, the relationship was not significant. The meta-regression method should be used to adjust the confounders in future study. Secondly, the target population of one study included in this meta-analysis was mainly on Caucasian population [20], but there was not enough data about Caucasian population provided by this paper. In our study, we used the information of all the population instead of the Caucasian population.
Despite some limitations, this meta-analysis indicated that subjects carrying at least one A allele of ERCC1 C8092A might increase the risk of glioma, whereas ERCC1 C118T polymorphism might have no influence on the susceptibility of glioma.
Acknowledgments
The authors have no support or funding to report.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
-
Chen J, McKay RM, Parada LF (2012) Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell 149: 36-47.
-
Jones C, Perryman L, Hargrave D (2012) Paediatric and adult malignant glioma: close relatives or distant cousins? Nat Rev Clin Oncol 9: 400-413.
-
Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M (2006) Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol 2: 494-503.
-
Kohler BA, Ward E, McCarthy BJ, Schymura MJ, Ries LA, et al. (2011) Annual report to the nation on the status of cancer, 1975-2007, featuring tumors of the brain and other nervous system. J Natl Cancer Inst 103: 714-736.
-
Marumoto T, Saya H (2012) Molecular biology of glioma. Adv Exp Med Biol 746: 2-11.
-
Melin B (2011) Genetic causes of glioma: new leads in the labyrinth. Curr Opin Oncol 23: 643-647.
-
Von Deimling A, Korshunov A, Hartmann C (2011) The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations. Brain Pathol 21: 74-87.
-
Goode EL, Ulrich CM, Potter JD (2002) Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 11: 1513-1530.
-
Ng PC, Henikoff S (2002) Accounting for human polymorphisms predicted to affect protein function. Genome Res 12: 436-446.
-
Ronen A, Glickman BW (2001) Human DNA repair genes. Environ Mol Mutagen 37: 241-283.
-
Zhu Y, Spitz MR, Amos CI, Lin J, Schabath MB, et al. (2004) An evolutionary perspective on single-nucleotide polymorphism screening in molecular cancer epidemiology. Cancer Res 64: 2251-2257.
-
Liang BC, Ross DA, Reed E (1995) Genomic copy number changes of DNA repair genes ERCC1 and ERCC2 in human gliomas. J Neurooncol 26: 17-23.
-
Smith JS, Tachibana I, Pohl U, Lee HK, Thanarajasingam U, et al. (2000) A transcript map of the chromosome 19q-arm glioma tumor suppressor region. Genomics 64: 44-50.
-
Wilson MD, Ruttan CC, Koop BF, Glickman BW (2001) ERCC1: a comparative genomic perspective. Environ Mol Mutagen 38: 209-215.
-
15. Matullo G, Guarrera S, Sacerdote C, Polidoro S, Davico L, et al. (2005) Polymorphisms/haplotypes in DNA repair genes and smoking: a bladder cancer case-control study. Cancer Epidemiol Biomarkers Prev 14: 2569-2578.
-
Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, et al. (2006) Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res 12: 2101-2108.
-
Zhou W, Liu G, Park S, Wang Z, Wain JC, et al. (2005) Gene-smoking interaction associations for the ERCC1 polymorphisms in the risk of lung cancer. Cancer Epidemiol Biomarkers Prev 14: 491-496.
-
Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, et al. (2006) Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 27: 560-567.
-
Liu Y, Scheurer ME, El-Zein R, Cao Y, Do KA, et al. (2009) Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol Biomarkers Prev 18: 204-214.
-
Wrensch M, Kelsey KT, Liu M, Moghadassi M, Sison JD, et al. (2005) ERCC1 and ERCC2 polymorphisms and adult glioma. Neuro Oncol 7: 495-507.
-
Chen P, Wiencke J, Aldape K, Kesler-Diaz A, Miike R, et al. (2000) Association of an ERCC1 polymorphism with adult-onset glioma. Cancer Epidemiol Biomarkers Prev 9: 843-847.
-
McKean-Cowdin R, Barnholtz-Sloan J, Inskip PD, Ruder AM, Butler M, et al. (2009) Associations between polymorphisms in DNA repair genes and glioblastoma. Cancer Epidemiol Biomarkers Prev 18: 1118-1126.
-
Woolf B (1955) On estimating the relation between blood group and disease. Ann Hum Genet 19: 251-253.
-
Lau J, Ioannidis JP, Schmid CH (1997) Quantitative synthesis in systematic reviews. Ann Intern Med 127: 820-826.
-
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177-188.
-
Tobias A (1999) Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull 8: 15-17.
-
Egger M SG, Altman DG (2001) Systematic reviews in health care: meta-analysis in context. 2nd (edn) BMJ London, UK.
-
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629-634.
-
Luo KQ, Mu SQ, Wu ZX, Shi YN, Peng JC (2013) Polymorphisms in DNA repair genes and risk of glioma and meningioma. Asian Pac J Cancer Prev 14: 449-452.
-
Pan WR, Li G, Guan JH (2013) Polymorphisms in DNA repair genes and susceptibility to glioma in a chinese population. Int J Mol Sci 14: 3314-3324.
-
Chen DQ, Yao DX, Zhao HY, Yan, SJ (2012) DNA repair gene ERCC1 and XPD polymorphisms predict glioma susceptibility and prognosis. Asian Pac J Cancer Prev 13: 2791-2794.
-
Zhang N, Lin LY, Zhu LL, Wu F, Wen H, et al. (2012) ERCC1 polymorphisms and risk of adult glioma in a Chinese population: a hospital-based case-control study. Cancer Invest 30: 199-202.
-
Martin LP, Hamilton TC, Schilder RJ (2008) Platinum resistance: the role of DNA repair pathways. Clin Cancer Res 14: 1291-1295.
-
Boiteux S, Gellon L, Guibourt N (2002) Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Radic Biol Med 32: 1244-1253.
-
Gellon L, Barbey R, Auffret van der Kemp P, Thomas D, Boiteux S (2001) Synergism between base excision repair, mediated by the DNA glycosylases Ntg1 and Ntg2, and nucleotide excision repair in the removal of oxidatively damaged DNA bases in Saccharomyces cerevisiae. Mol Genet Genomics 265: 1087-1096.
-
Zhang Z, Zhou C, Zhang J, Tang L, Su B (2008) Relationship between polymorphisms of DNA repair gene ERCC1 and susceptibility to lung cancer. Zhongguo Fei Ai Za Zhi 11: 183-188.