Osteoarthritis (OA) is among the most prevalent joint diseases. Reduced patient life quality and productivity represent a major personal and community strains. GDF5 gene is involved in the development of bone and cartilage. Studies reported a clear and highly reproducible association between susceptibility of knee osteoarthritis (KOA) and GDF5. This study aims to detect GDF5 gene polymorphism and to evaluate its association with susceptibility to KOA.
This study is a case-control carried out at the female section of College of Applied Medical Sciences, Taif University. 50 Samples with KOA were collected from patients at King Faisal Medical Complex (Rheumatology Clinic), Taif, KSA. The body mass index (BMI) and the functional disability were estimated in all patients. CRP and RF were estimated in the serum of all included subjects by ELISA. ESR was determined by Westergren method. GDF5 T > C (rs143383) promoter polymorphism was assessed by PCR- RFLP.
We observed a statistically significant elevation of CRP, RF, and ESR on the patients with KOA relative to the controls. Seventy-two percent of our patients have obesity grade I and II and 56% have severe functional disability on Womac index. The TT and TC genotypes of GDF5 gene polymorphism were statistically more frequent in KOA patients than in the controls. TT genotype may be a risk factor to KOA. The T allele was more frequent in KOA and C allele was more frequent in the controls. GDF5 polymorphism was not related to BMI and functional disability of KOA. GDF5 polymorphism was found to be significantly related to high ESR and CRP.
The present study revealed a possible genetic link between KOA and GDF5 polymorphism and that the TT genotype may increase the risk of development of KOA. However, a future study with a large number of patients is needed to confirm our results.
Knee osteoarthritis (KOA), Gene polymorphism, Growth differentiation Factor 5 (GDF5)
OA: Osteoarthritis; GDF5: Growth Differentiation Factor 5; BMP: Bone Morphogenetic Protein Group; TGF-β: Transforming Growth Factor-β; BMPR-IB: BMP Receptor IB; ActR- Type IIB: Activin Receptor (ActR) Type IIB; SNP: Single Nucleotide Polymorphism; 5'UTR: 5' Untranslated Region; GWAS: Genome-Wide Association; CRP: C Reactive Protein; KOA: Patients with Knee Osteoarthritis; ESR: Erythrocyte Sedimentation Rate; RF: Rheumatoid Factor; BMI: Body Mass Index; WOMAC: The Western Ontario And Mcmaster Universities
Osteoarthritis (OA) is a widespread polygenic multifactorial disorder accompanied by joint cartilage degeneration and synovial inflammatory changes [1]. OA is the primary cause of physical disability in the elderly, and is characterized by pain in the joints, inflammation, and rigidity [2,3]. It is among the most prevalent joint diseases [4]; the incidence of hip and/or knee OA with symptoms is about ~ 242 million worldwide [5], with other disorders it is Slisted as the eleventh highest international disability cause [4]. Reduced patient life quality and productivity represent a major personal and community strain with a universal occurrence of 3.8% [6,7].
Knee OA has a higher prevalence than other joints, with an incidence rate of approximately 45 percent, rising to 60.5 percent among obese individuals [8]. Incidence rises with each decade of age and with the highest annual occurrence between ages 55-65 [9]. Knee OA is considered to be a widespread joint degenerative disorder primarily affecting the elderly [10]. It is characterized by destruction of the cartilage of joints [11]. Precise etiology for OA evolution remains lacking. However, age, genetic factors, environmental conditions and inappropriate lifestyles are the known threats for OA [12]. The effect of certain genes is examined, as the genetic influence of the primary OA is high, i.e. 40% for the knee, 60% for the hip and 65% for the hand joints [13]. Growth differentiation factor 5 (GDF5) gene is located on the chromosome 20q11.2 and it regulates the expression of GDF5 protein. It is classified within the bone morphogenetic protein group (BMP) [14]. It involves in development of bone and cartilage, particularly in endochondral ossification process [15]. The mutation of GDF5 gene associates with generalized osteoarthritis and skeletal related congenital diseases [16].
GDF5 is recognized also as the cartilage-derived morphogenetic protein of the superfamily transforming growth factor-β (TGF-β), have been found to play a key role in the growth, repair and reconstruction of cartilage and bone [17]. GDF-5 binds to the transmembrane serine/threonine kinase type I and II receptors to activate its signaling pathway [18]. BMPR-IB (BMP receptor IB), BMPR-II, and activin receptor (ActR) type IIB have higher affinities for the GDF-5 ligand [19]. Upon GDF-5 binding, the receptors are phosphorylated to activate the downstream Smad pathway. The Smad proteins then translocate into the nucleus to regulate the transcription of various genes [20].
Several studies have revealed that GDF5 performs a major role in musculoskeletal processes, influencing endochondral calcification, joint development, tendon repair and bone formation [17]. Defects of this gene were shown to be correlated to abnormal joint development or skeletal disorders in humans and mice [21]. Moreover, the polymorphism in GDF5 gene is related with low expression of the GDF5 protein in knee joint [22]. In European and Asian populations, rs143383 polymorphism in the 5'-untranslated regions of GDF5 is associated with knee OA [23]. In this regard, studies on OA-associated single nucleotide polymorphism (SNP) rs143383 is a C to T transition located within the 5' untranslated region (5'UTR) of GDF5, which codes for the extracellular signaling molecule GDF5. About the association between rs143383 and OA, the investigators demonstrated that the OA-risk T-allele of the SNP-mediated reduced mRNA expression relative to the C-allele in an luciferase assay conducted in a chondrocyte cell line; chondrocytes are the only cell type present in cartilage [22]. This study stressed rs143383 as the vital functional SNP responsible for OA. It was also shown that the T-allele was associated with diminished production of GDF5 in cartilage [17].
Studies of genome-wide association (GWAS) reported a clear and highly reproducible association between susceptibility of knee OA and GDF5. In particular, single nucleotide polymorphisms (SNPs) span a length of 130 kb comprising GDF5 and downstream UQCC1 (ubiquinol-cytochrome C reductase complex assembly factor 1) was related with a 1.2 to 1.8-fold raise in knee OA risk [24].
Genetic factors are known to be important in the pathogenesis of osteoarthritis. The etiology of osteoarthritis remains complex, and the latest genetic information indicates that susceptibility to osteoarthritis occurs from unique combinations of multiple gene-gene and gene-environment interactions. This study was carried out to detect GDF5 gene polymorphism and to evaluate its association with susceptibility to knee osteoarthritis.
This study is a case-control conducted in the female section at College of Applied Medical Sciences, Taif University. The study has been accepted by the ethical board of Collage of Applied Medical Sciences, Taif University and all participants gave informed consent to participate in the research.We included 50 patients with Knee OA (13 males and 37 females, their age ranged between 25 and 66 years) who attended in King Faisal Medical Complex (Rheumatology Clinic), Taif, KSA. Fifty healthy individuals with matched age and sex with the Knee OA patients and with no clinical and laboratory evidences of OA or other diseases were also included as a control group. Inclusion criteria include all patients with KOA were diagnosed according American College of Rheumatology [25]. Antero-posterior and lateral view of knee radiographs were taken to confirm the diagnosis of OA by Kellgren and Lawrence scores [26]. Exclusion criteria include patients with other rheumatologial diseases such as: Systemic lupus erythematosus, mixed connective tissue diseases, Scleroderma, and Dermatomyositis.
The BMI was estimated for each patient using the equation (BMI = Weight kg/Height m2). Functional disability in OA was assessed in all patients according to Kellgren score [26], the Western ontario and McMaster universities osteoarthritis index (WOMAC) [27], and the Lequesne index [28].
A 10 ml peripheral venous blood sample was collected from all included subjects. Five milliliters of each obtained sample was withdrawn in EDTA tubes for DNA extraction and ESR determination. The remaining sample was left for 1 hour in a serum separator collection tube to clot at room temperature and centrifuged at 3000 rpm for 5 minutes. The separated serum was obtained and stored in -20 ℃ until analysis. CRP and RF levels were estimated in serum samples from KOA patients and control subjects using Human C Reactive Protein ELISA Kit (CRP), abcam, Cambridge, MA, USA (Cat No. ab99995), and rheumatoid factor (RF), ELISA Kit, MyBioSource, Inc. San Diego, CA 92195-3308, USA (Cat No. MBS262327) respectively. ESR was determined using Westergren method.
Genomic DNA was extracted and purified using the QIAamp DNA mini kit (Qiagen CA, USA) from the peripheral blood collected in tubes containing EDTA from knee osteoarthritis patients and controls. Purified DNA was kept at -80 ℃ until used for genotyping.
Genotyping of GDF5 T > C was performed using the polymerase chain reaction restriction fragment length polymorphism (PCR- RFLP) as described by Tulyapruek, et al. [29]. The PCR amplification was carried out using recombinant Taq polymerase master mix (Dream taqgreen, code number k1081, LOT: 00643300) (Thermo Fisher Scientific Ballics UAB, V A Cracino 8, LT-002241 Vilnius, Lithuania) in a 25 µl total volume. Primer sequences to amplify the promoter (rs143383) of GDF5 were: GATTTTTTCTGAGCACCTGCAGG (forward) and GTGTGTGTTTGTATCCAG (reverse). Initial denaturation for 5 minutes at 95 ℃ followed by amplification for 35 cycles in thermocycler (PCR Sprint, Thermofisher, Waltham, MA) and denaturation for 1 minute at 94 ℃, then annealing for another 1 minute at 58 ℃, extension for 1 minute at 72 ℃ and final extension for 10 minutes at 72 ℃. Following the manufacturer's standard, 10 µL of PCR sample was incubated with 3 units of BsiEI restriction enzyme for 4 hours at 37 ℃. The digested product was electrophoresed on 2% agarose gel with ethidium bromide staining before being visualized on a UV transilluminator. The fragments lengths were 104 and 230 bp in CC, 104, 230, and 344 bp in TC, and 344 bp in TT genotypes.
Using the SPSS computer software version 22.0., the data was recorded, tabulated and analysied. This research showed qualitative data as mean and standard deviation, with a number and a percentage, and quantitative data. Chi-square (X2) test was used for comparing qualitative variables. The value of P < 0.05, and Odd ratio (OR) and Confidence interval (C.I.) of 95% are taken as important.
The present study included 50 patients with knee osteoarthritis (KOA) and 50 healthy individuals. The age of the patients ranged 25 and 66 years while in the controls it ranged between 28 and 68 years. It included 13 male patients and 37 females and14 male and 36 females of healthy individuals, Table 1.
Table 1: Age and sex of included subjects. View Table 1
The C-reactive protein level (CRP) in KOA patients ranged between 1.01 and 8.12 mg/L with a mean value ± SD of 4.324 ± 2.045 while in the controls it ranged between 0.004 and 2.03 with a mean ± SD of 0.91 ± 0.5. The difference of CRP levels in patients and controls was statistically significant (t = 11.48, P < 0.0001), (Figure 1A). In patients with KOA the ESR ranged between 3 and 20 mm/hr with a mean ± SD value of 10.48 ± 3.52 mm/hr. In control individuals ESR ranged between 1 and 13 mm/h with 7.68 ± 2.97 mm/h as a mean ± SD value. The difference between RA patients and the controls was statistically significant (t = 4.299, P < 0.0001), (Figure 1B). The RF was assessed in the serum patients with KOA and the controls, in patients the serum level ranged between 3.45 and 12.24 IU/ml with a mean ± SD value of 6.5 ± 2.7 IU/ml while in the controls it ranged between 0.023 and 5.16 with a mean ± SD value of 2.78 ± 1.4 IU/ml. The difference between both groups was statistically significant (t = 8.73, P < 0.0001), (Figure 1C).
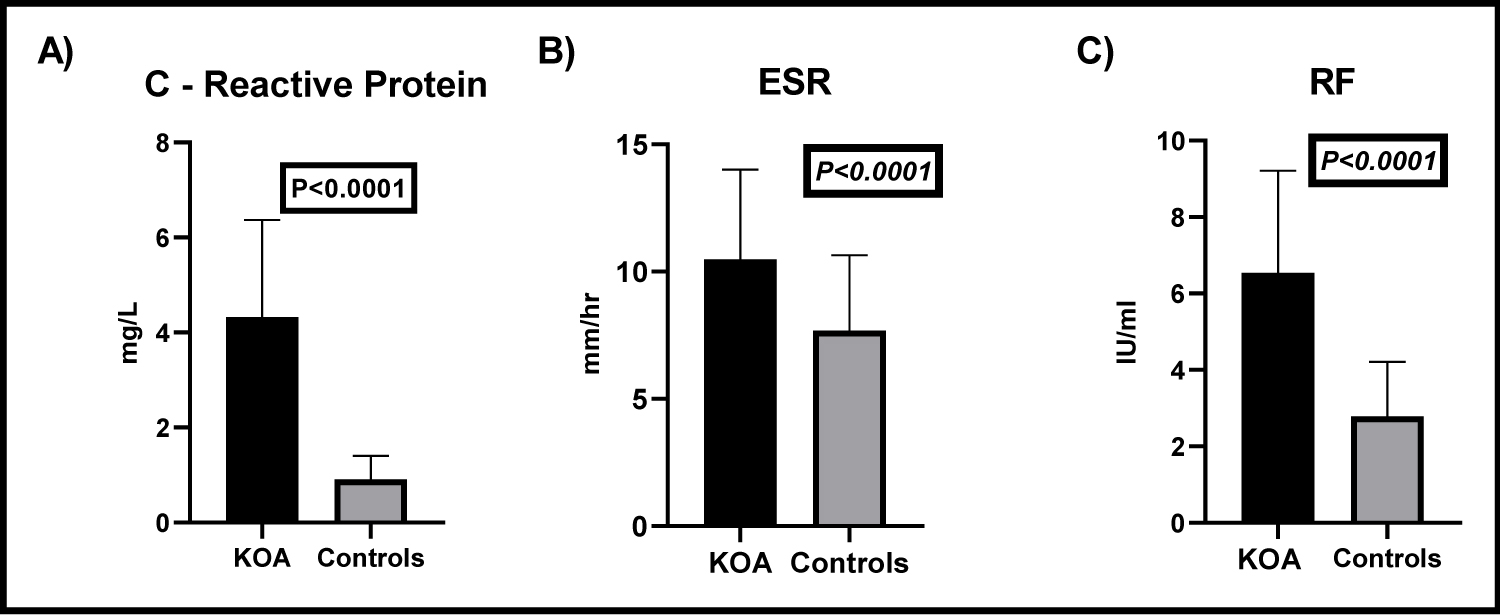 Figure 1: Estimation of the levels of CRP, ESR, and RF in patients with KOA and controls.
Figure 1: Estimation of the levels of CRP, ESR, and RF in patients with KOA and controls.
A) CRP levels in patients with KOA and controls. CRP levels in KOA patients were significantly increased than in controls (t = 11.48, P < 0.0001); B) Erythrocyte sedimentation rates of both KOA patients and the controls. ESR in KOA patients was significantly increased than controls (t = 4.299, P < 0.0001); C) Levels of RF in the sera of KOA patients and controls. The RF was significantly elevated in the serum of patients with KOA, compared to the controls (t = 8.73, P < 0.0001).
View Figure 1
Assessment of BMI in the included patients of KOA showed that most of our included patients were suffering from obesity, Figure 2.
 Figure 2: Body Mass Index of all included patients with KOA.
View Figure 2
Figure 2: Body Mass Index of all included patients with KOA.
View Figure 2
We analyzed the functional disability in patients with KOA by the use of the most common indices available. According to the kellgren score, 44% of our patients have grade II, 36% grade I, 14% with grade III, and 6% was classified as grade 0. Using the Lequesne index, most of our included patients about 34% have a mild disability while extremely severe disability was recorded in 6% of studied patients. The Western ontario and McMaster universities (WOMAC) Osteoarthritis Index, depends on the assessment of pain, stiffness, and function. Sixty-five percent of our studied patients have a more severe WOMAC score, Table 2.
Table 2: Functional disabilities in KOA patients. View Table 2
We analyzed the frequency of GDP5 gene polymorphism in KOA patients; we observed that TT and TC genotypes were more frequent in KOA than in the controls and that TT genotype may be a risk factor to OA. The difference in the frequency of GDP5 polymorphism genotypes were statistically significant between KOA patients and the controls (P = 0.019), Table 3.
Table 3: GDF5 genotypes polymorphism in the studied groups. View Table 3
Analysis of alleles frequency of GDF5 gene polymorphism revealed that the T allele was more frequent in KOA (56%) and C allele was more in the controls (62%). This difference was statistically significant P = 0.006, Table 4.
Table 4: Alleles frequency of GDF5 polymorphism in the studied groups. View Table 4
There was no statistically significant correlation between BMI and GDP5 polymorphism in our included patients (X2 = 1.24, P = 0.99), Table 5.
Table 5: Relation between GDP5 gene polymorphism and BMI in patients with OA. View Table 5
Table 6: GDP5 genotypes and different studied laboratory parameters. View Table 6
Table 7: GDF5 genotypes and functional disability in KOA. View Table 7
Assessment of the frequency of different genotypes of GDP5 polymorphism in relation to the presence of anemia in KOA patients revealed no significant relation, while there was a significant relation between high ESR, high CRP and GDP5 genotypes, P = 0.045 and 0.033 respectively, Table 6.
Assessment of the functional disability in patients with OA according to the different GDP5 genotypes revealed no significant relation between genotypes and Kellgren score (P = 0.715), Lesquene index (P = 0.79), and Womac OA index (P = 0.65), Table 7.
Osteoarthritis (OA) is the most common chronic, degenerative, and disabling joint disease worldwide [30]. Primary knee OA is the most prevalent type of OA that usually affects people over age 45. Knee OA contributes to functional and psychological and social dysfunction related to quality of life impairment [4]. The molecular history of primary knee OA includes several genes encoding proteins that have important roles in the mechanism of underlying disease. Prior researches had examined many OA-associated target genes in various communities [7]. However, the results of most reports did not achieve a consensus on the identified OA susceptibility gene. The genes involved in the pathogenesis of OA are not clear at the moment [15]. Many studies have suggested that mutations of the GDF5 gene can lead to this disorder but the findings of genetic correlation research have been conflicting due to the feasibility of reproducing important correlations [17].
GDF5 is a signaling molecule, which is involved in development of the bone and cartilage, as well as joint formation [15]. Genetic abnormalities in the GDF5 gene develop a wide spectrum of masculo-skeletal diseases [31]. In the current study, we observed that GDF5 gene polymorphism was significantly related to OA of the knee. The current work revealed that there was a significant difference in the frequency distribution of the GDF5gene polymorphism between KOA patients and controls. The TT genotype polymorphism was more frequent in KOA relative to controls. Also, the T allele was common in KOA. Our results support the results of several previous studies who also noted in their study this significant correlation [22,28,29,32]. On the contrary, several studies reported that there is no relation between GDF5 polymorphism and KOA [17,30,31].
Many reasons may be related to the variation of results between the studies published worldwide [32,33]. First it is widely recognized that gene and environment and their interactions are influencing the development of OA and all previous studies were carried out on different populations with different genetics profiles. Second, variation in patient’s recruitment criteria and methods of genotyping may result in variation of results between studies. Third, it has shown also that OA is a multifactorial disorder [32]. In addition to the above mentioned factors another GDF5 polymorphism in the same region (rs143384) may affect the GDF5 rs143383 expression; moreover, several different polymorphisms, separate from rs143383, which affect the GDF5 allelic expression were observed [33-38].
In the present study, we also studied the genetic influence of GDP5 polymorphism and different laboratory parameters in KOA. We found a statistically significant correlation between GDF5 polymorphism especially TC genotype and high ESR and high level of CRP. Zhang, et al. was also previously reported that a significant correlation between TC genotype and high ESR and with high CRP [33]. In addition, the present study, there was no significant correlation between GDF5polymorphism and body mass index (BMI) in patients with KOA. Our results support the results of Mohasseb, et al. [34]. On the contrary to our results Zhang, et al. who reported that a significant correlation between GDF5 polymorphism and increased body mass index [33]. We analyzed the GDF5 polymorphism with the functional disability in patients with KOA. We noted that there is no significant correlation between GDF5 polymorphism and functional disability in KOA. Our results agree with the previous results reported by Zhang, et al. [33]. On the contrary, Mohasseb, et al. reported a significant correlation between functional disability in KOA and GDF5 polymorphism [34]. Collectivity, the current study revealed a possible genetic link between KOA and GDF5 gene polymorphism (rs143383) and that TT genotype may increase the risk of development of KOA. Our results need to be confirmed by a study with large number of patients.
Not applicable.
Authors declared no conflict of interest.